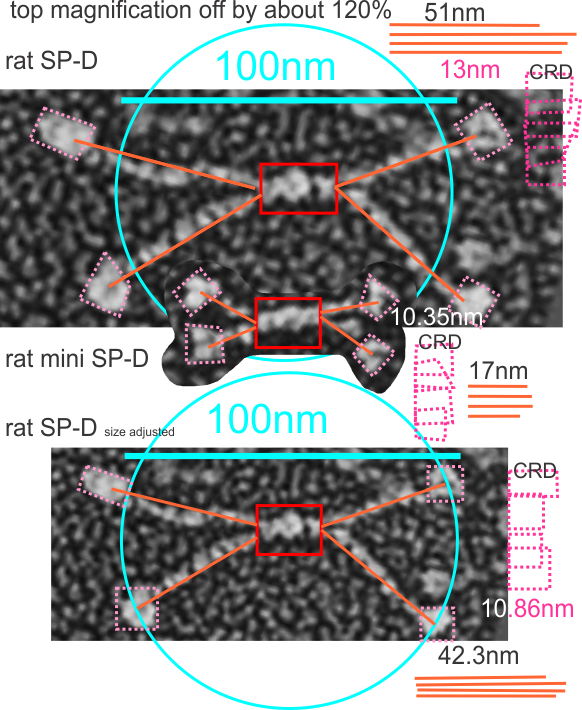
The publication by White et al, mentioned in previous posts shows rat SP-D as well as a two genetically modified SP-D proteins (as dodecamers) which I am trying to analyze to estimate changes in arm length on micrographs after the deletion of exons (they referred to as C 3 and C 4 in the collagen like domain) from SP-D. Adjusting their original bar marker for magnification (this i adjusted on the basis that the CRD elements in the mini SP-D (abiyt 10.35nm) and the rat SP-D should be the “same or very similar” in size, and theirs was about 13nm which is 115 to 120% larger in the SP-D than the mini-SP-D which i don’t think it should have been. And also the background granules in the three panels were not the same, and were larger in their top image than in B and C, which seemed strange. Both measurements are given, that is, the difference between their original micrograph and the slightly reduced rat SP_D micrograph (so that the SP-D dodecamer is more along the size lines of what has been reported by countless authors before this particular publication) i.e. just over 100nm.
Also the relative differences in size of the “arms” of the dodecamers of the mini SP-D are given. the miniSP-D is about 33.3 percent of the length of the original SP-D dodecamer arm. I also look like the length measure will not differ from the arc length when measured using that dimension. IF you use their magnification marker for all images. If you use the size of SP-D adjusted so that the CRD are quite similar to the CRD in the mini SP-D, then the mini-SP-D arms are about % of the adjusted SP-D size. The mini SP-D and lower image of SP-D are relatively similar magnifications (at least as measured by their CRD.