I redid this same image of mini-surfactant protein D, and made sure that the straight lines and the arc length lines were pretty closely spaced. This is a bit of a moral problem since I don’t think the exact location of the end of the collagen-like sequence, or even the beginning of that sequence are known. Especially in the center area of the molecule (both the rat unmodified SP-D or the mini, or human or whatever) the length of tethering in the SP-D dodecamers appears to be much greater (upwards of 25% of the total length of the mini-SP-D) than the number of “counted” amino acids for the neck and N terminal. The neck and CRD are modeled on RCSB as a unit, with the coiled coil neck not being exposed very much from the CRD to which it attaches –almost folded back upon.
There could be some close associations in the C1 exon of the SP-D collagen like domain that measuring from the “bifurcation” seen in the dodecamers does not measure. It seems likely that it is in the middle part of the collagen like domain because this measurement does not appear to have changed at all in the mini-SP-D. But below is a repeat … and just my opinion, it isn’t going to make (at least with the mini-SP-D there is not much difference if arc length or straight length for the dodecamer arm is used for the measurement. Clearly one nm difference in the length estimated by these measures is smaller than the bias in the line drawings). I think it is pretty safe to say that mini-SP-D is about 70nm in diameter (that would be the diameter (70 nm) of a circle which intersects at least two of the CRD. Because of the similarity in measurements i will not do arc length for the mini-SP-D.
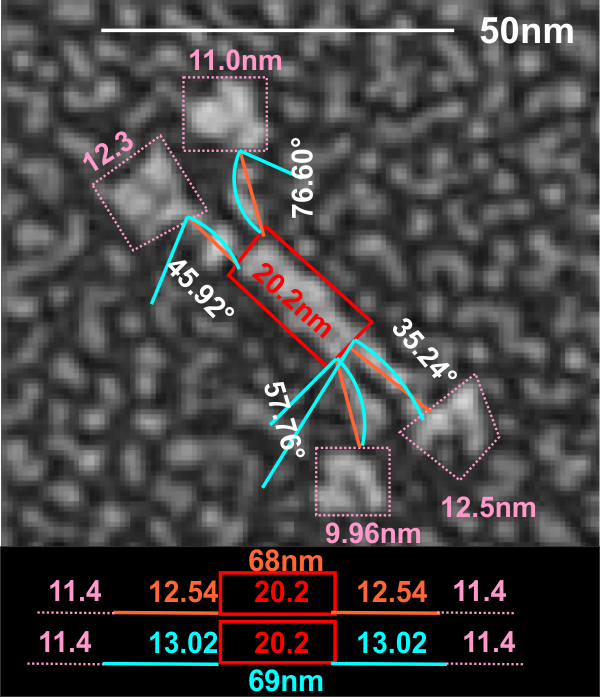 This measure of 68 or 69 nm for mini SP-D makes it about 60% the size of the full sequence SP-D. Because the measures of the tethered portion of the center of the dodecamer and the CRD are so close in measures, one can assume that the reduction was somewhere in the middle of the collagen like sequence.
This measure of 68 or 69 nm for mini SP-D makes it about 60% the size of the full sequence SP-D. Because the measures of the tethered portion of the center of the dodecamer and the CRD are so close in measures, one can assume that the reduction was somewhere in the middle of the collagen like sequence.