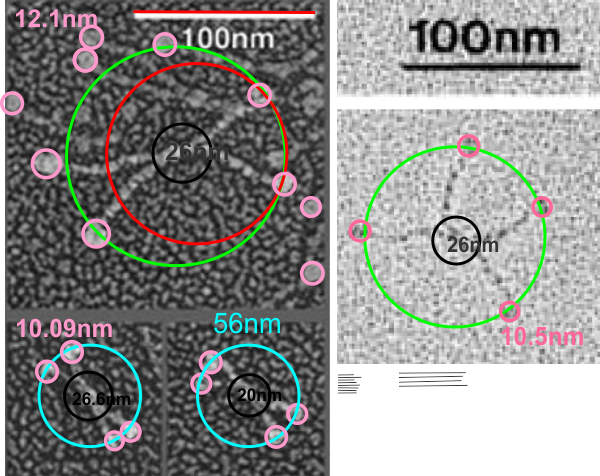
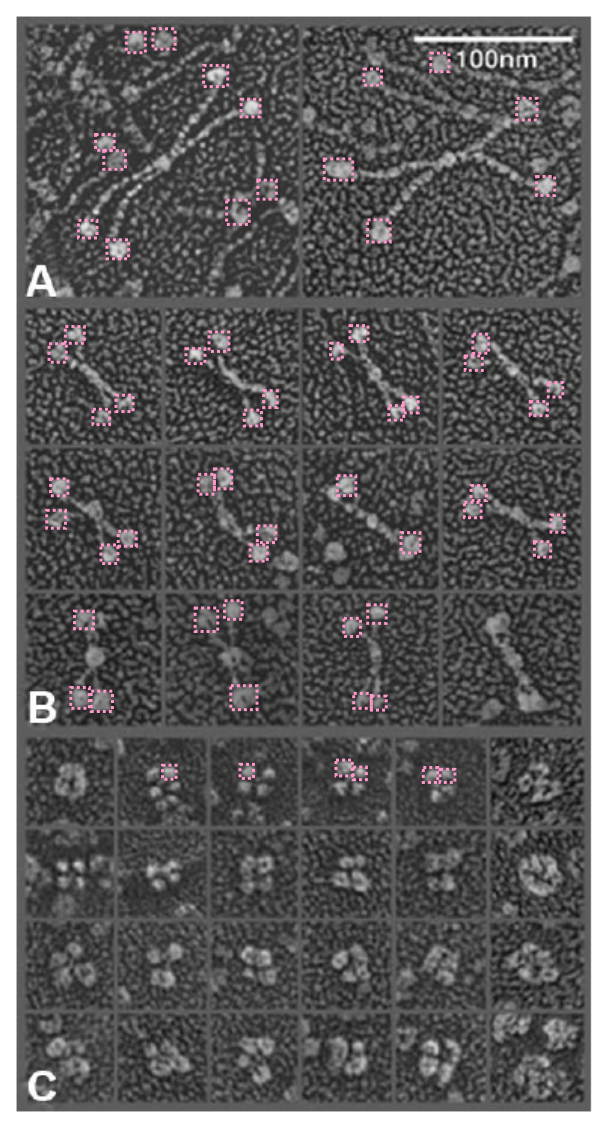
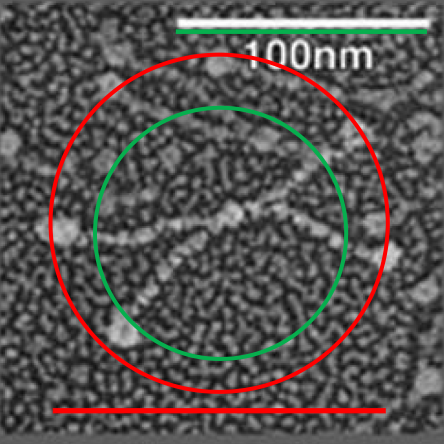
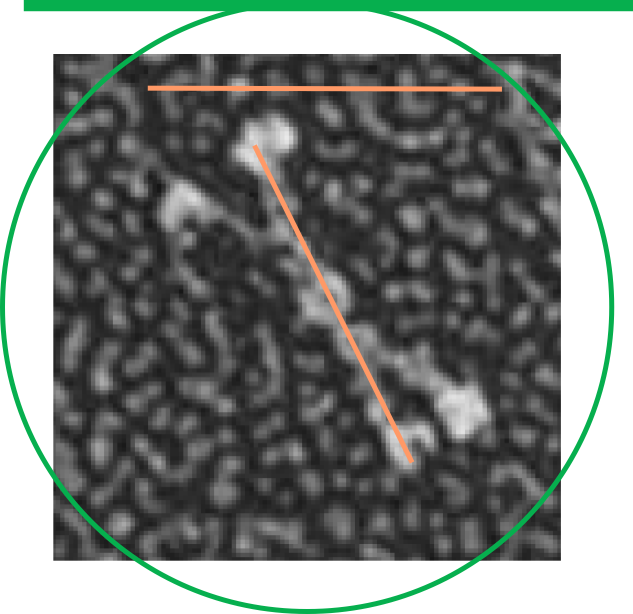
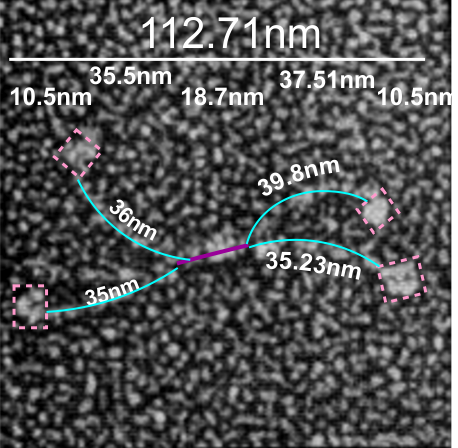
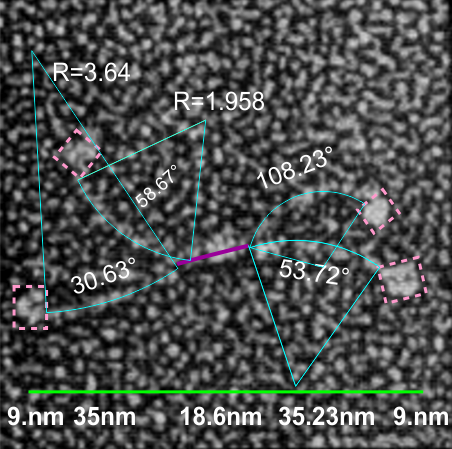
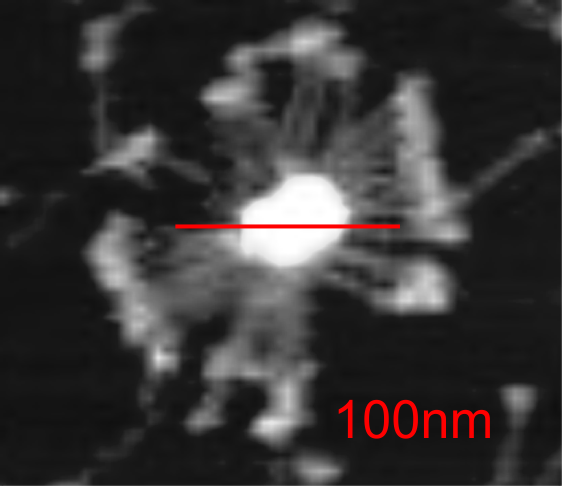
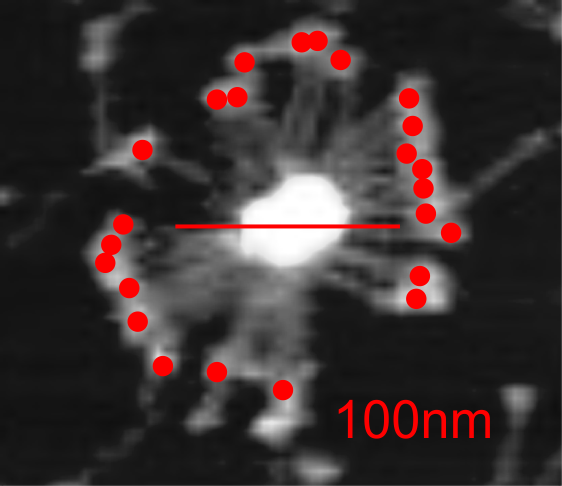
It is pretty obvious that the center of SP-D (at least as is represented in dozens and dozens of TEMs, and AFMs ) is longer in the direction of the 45 degree angle than the amino acid sequence would suggest and longer too than most diagrams present. There is a central dense region (which is found also on the tracing) but an extended region where two arms of the dodecamer are closely aligned with the combined length of something around 30nm, or a little less than a third of the entire length of the dodecamer. The separation of the two arms on either end into the @45 degree angle leaves the remainder of the arms as the collagen-like domain, and the neck and CRD on the order of 10-13nm the latter folded back upon the neck section to some degree. Tracing from Figure 2. Arroyo et al. JMB. 430, 1495-1509, 2018. Their measurement for the total width of a surfactant dodecamer seemed to be considerably larger than measured by others. So a compromise in estimated dimension has to be taken because of the differences in measurement accuracy and how much spreading of the molecules has occurred… Relative measurements then, of 100 nm might be useful in determining the relative lengths of the segments of the dodecamer, but not the absolute lengths.
Normalizing the dodecamers to 100nm cutting centrally through at least two (probably best making it two polar) CRD is useful. A summary of the center (the highest, brightest (by TEM and AFM) areas of the SP-D dodecamer might be about


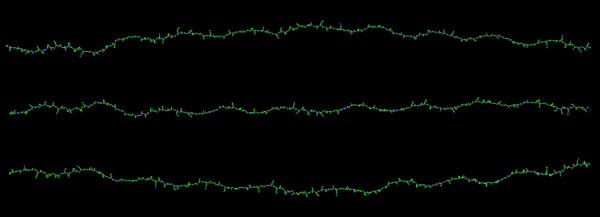
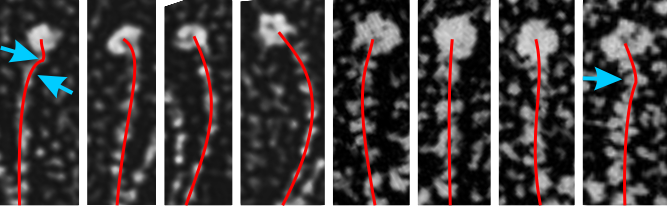
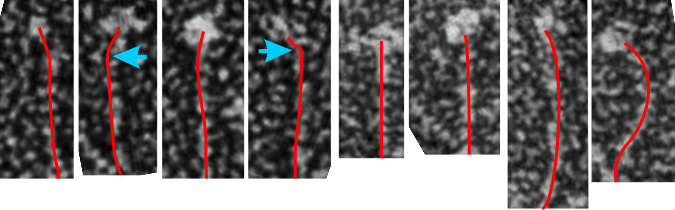
I would like to find a way to trace density of a curved line…. straight lines are no problem. I think it is time to go back and measure these all one more time but setting density and width standards to get relative dimensions which at some point could be used for generalities describing proportions and regions involved in SP-D. s