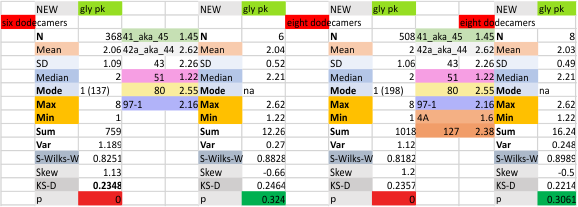
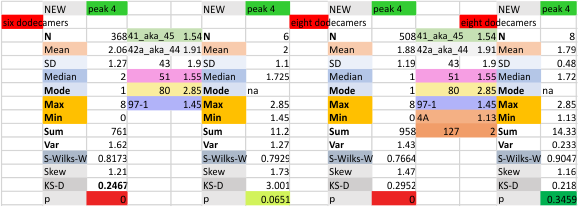
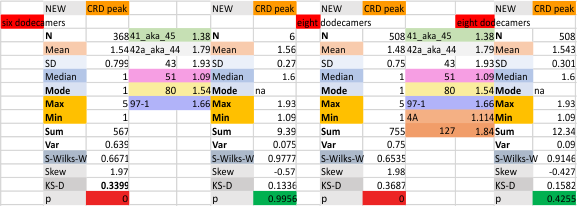
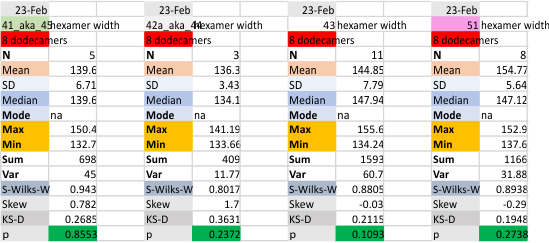
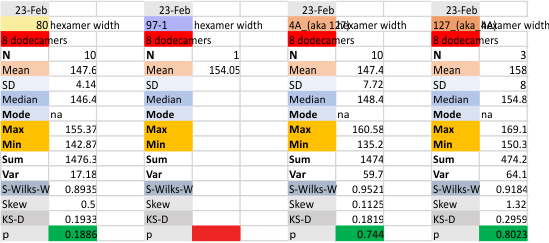
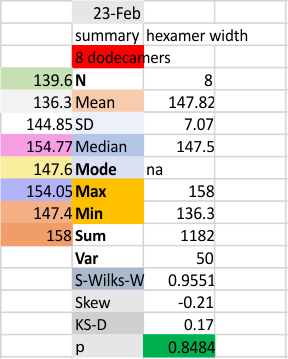
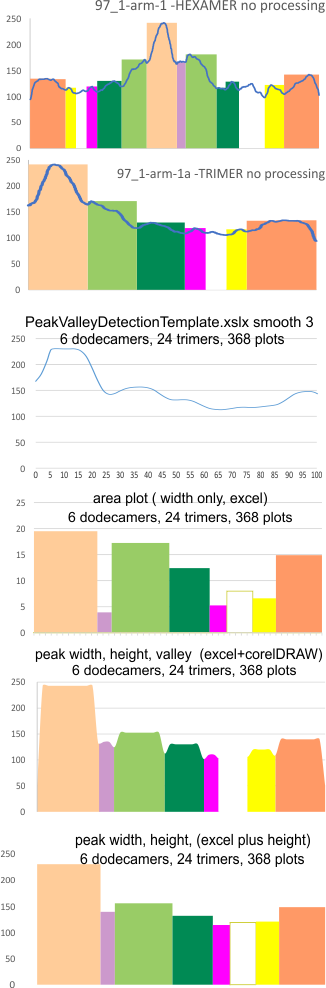
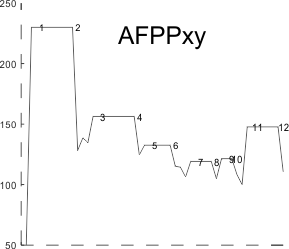
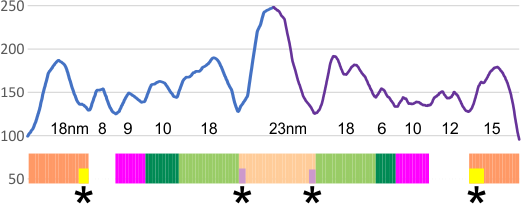
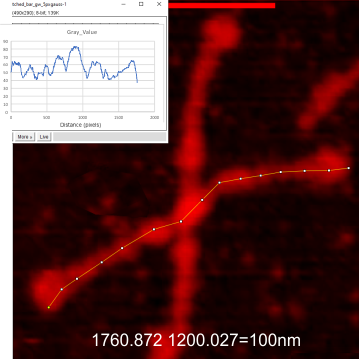
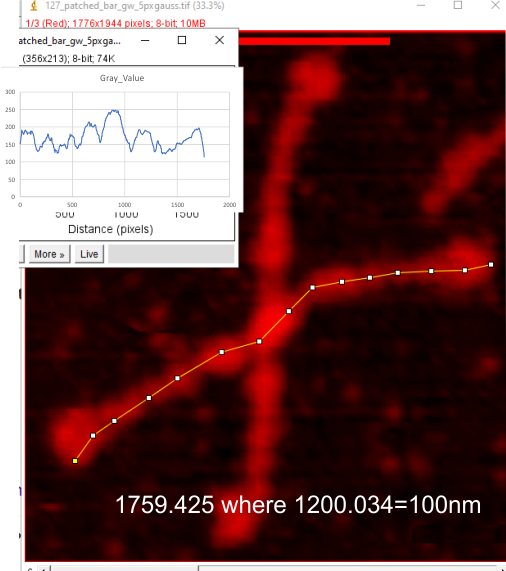
These data show the number of smaller peaks within the tracings of SP-D trimers (using AFM images from various published papers). At this point they are all rhSP-D images. The trimers are plotted beginning at the most complete side of the N term peak. This means that the whole N term is plotted (and it includes the N from the adjacent trimer(s) that build a dodecamer. All these plots are from dodecamers. This link is to a summary of the number of sub-peaks.
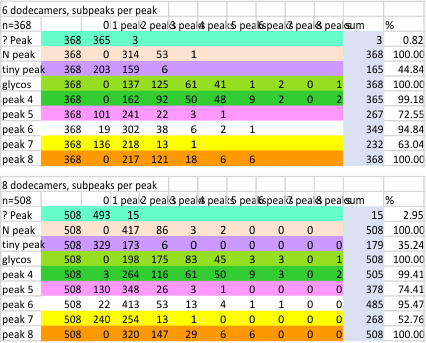
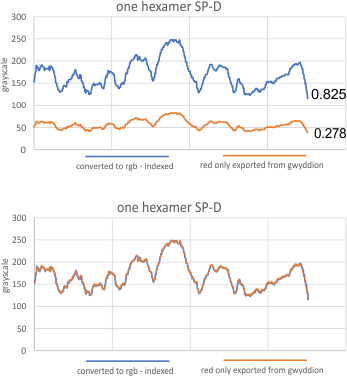
It has been my observation that the multimers of SP-D with higher than four trimer arms often show a decreased brightness within the center, and many images confirm this. In this series of plots there are just a few images where in a plot of a hexamer, there is a small peak in the center of the N term peak. I have labeled it the ? peak, and it is shown below in light bluegreen. It doesn’t occur often but enough to mention it. All the data are organized similarly. On the far left is the sum total of peaks from six hexamers (n of trimers =368 which includes gray scale plots from many signal and image processed images, so on the very left, no division into dodecamers is made, but the column just to the right of that has six dodecamers where the mean occurrence of peaks is found where the N=6. On the right hand side of the data, far right, the same has been applied to 8 dodecamers, and just to the left of that set of numbers is data for each individual trimer, and n=508 plots.
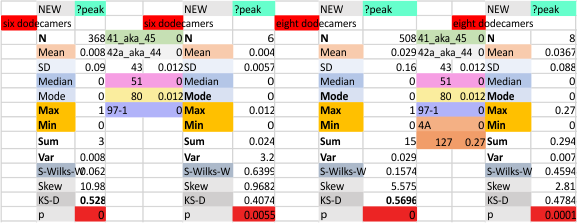 While signal processing apps have determined that there are likely 15 peaks per hexamer, this number does NOT include the ? peak above.
While signal processing apps have determined that there are likely 15 peaks per hexamer, this number does NOT include the ? peak above.
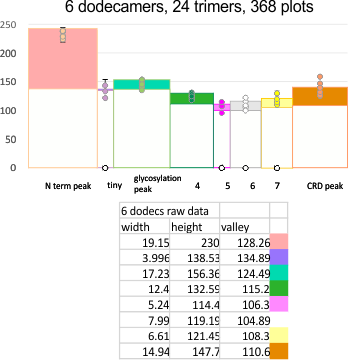
The 8 peaks per trimer (N peak is counted once with each trimer, but also counted only once with each hexamer) are as follows:
N term peak, tiny unidentified peak, glycosylation peak, peak 4, peak 5, peak 6, coiled neck peak 7, CRD peak 8, and are labeled like this below. The means are the number of sub-peaks per peak. The three confirmed peaks (in the literature are N, gly and CRD, the neck peak 7 is inconsistent because the CRD domains obstruct it often. Peaks that are NEW, and as yet unconfirmed are tiny peak (2), peak 4, 5, and 6).
The N term peak has been organized with the mean of the sub-peaks per N term peak plotted from the same trimers as above. And so on, for each of the peaks — as listed above.
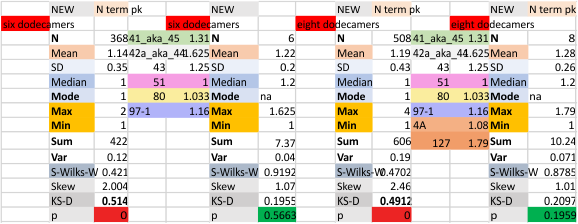
The glycosylation peak is apt to be not just one peak but two (and my thought is because each of the trimers can by glycosylated individually, and the coil of the trimer ofsets each of those sub-peaks. Those peaks composed of more than one sub-peak are the glycosylation peak (peak 2) and an as yet undescribed peak 4 which also has two sub-peaks. The CRD domain is subject to a different type of flopping around, and does not show two peaks consistently. This is likely affected by the fact that a plot line goes through the CRD often not picking up the apparently random order that the CRD domains of the trimer fall during processing.