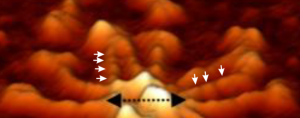
Arroyo’s publication on surfactant protein D certainly my favorite, and the supplemental figures shows images which confirm pretty clearly (in my own mind anyway) that there are 3 or 4 peaks between the N termini connections and the CRD. See below a portion of her image which i have cropped and added white arrows to pointing out how the peaks show up both in a lateral view and foreshortened view. Her figure showing the central diameter of the N termini and part of the collagen-like-domain is the black dotted line with black arrowheads.
This is a crop from her supplemental figure.
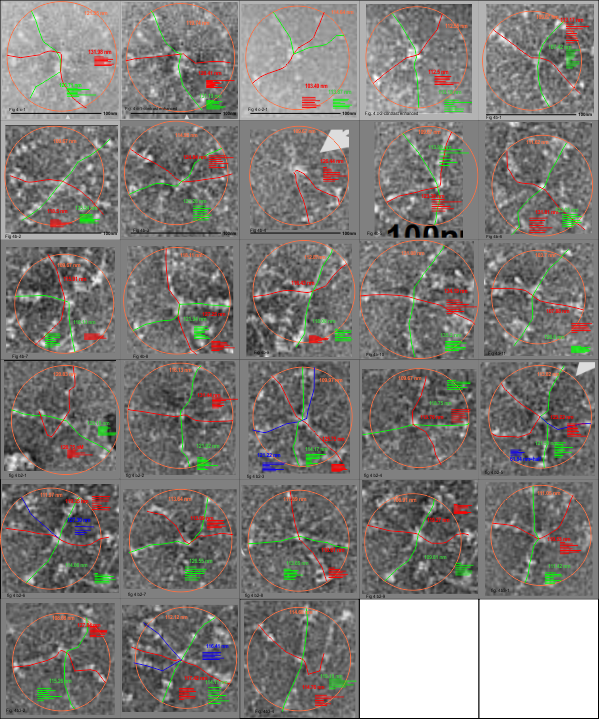
I am glad to have the supplemental figures to check out the size of the SP-D dodecamers and their brightness (LUT) plots and arm length using vector and node lines) and straightened molecules I have made with the cover images, against one of the supplemental figures which appears to be a similar area just rotated and cropped but actually has a micron bar marker (unlike the cover image legend which was gave the size in text, erroneously).
Monthly Archives: March 2020
Two methods for measuring SP-D
1-tailed t-test for seeing if measures of arm length spanning a hexamer, that is from the outer edges of one CRD through the N terminal junctions and out another arm to the other CRD. of a diameter (touching 3 of the four CRD trimers at the ends of at least 3 of the four arms of SP-D (the smallest diameter able to be made).
Previous t-test has not shown a difference, however using all measures to date which includes AFM, rotary shadowed and negative stained images (all available in published documents) there may be a significant difference between methods. Below n=291 arms (which is 145 dodecamers) using the two methods (where n=diameter is
half as many as the separate measures of two hexameric arms.
Difference Scores Calculations
Two hexamers measured with a vector/node line running from CRD to CRD)
N1: 291
df1 = N – 1 = 291 – 1 = 290
M1: 133.8
SS1: 187767.45
s21 = SS1/(N – 1) = 187767.45/(291-1) = 647.47
One measurement of the smallest diameter which touches the outer edge of three of the four CRD.
N2: 146
df2 = N – 1 = 146 – 1 = 145
M2: 127.48
SS2: 82472.16
s22 = SS2/(N – 1) = 82472.16/(146-1) = 568.77
T-value Calculation one-tailed
s2p = ((df1/(df1 + df2)) * s21) + ((df2/(df2 + df2)) * s22) = ((290/435) * 647.47) + ((145/435) * 568.77) = 621.24
s2M1 = s2p/N1 = 621.24/291 = 2.13
s2M2 = s2p/N2 = 621.24/146 = 4.26
t = (M1 – M2)/√(s2M1 + s2M2) = 6.32/√6.39 = 2.5
The 1-tailed t-value is 2.49859. The p-value is .006418. The result is significant at p < .05.
The 2-tailed t-value is 2.49859. The p-value is .012837. The result is significant at p < .05
Values from the two measurement approaches when the Arroyo mislabeled nm cover image measurements are removed from each type of measurement
Difference Scores Calculations
Two hexamers measured with a vector/node line running from CRD to CRD)
N1: 261
df1 = N - 1 = 261 - 1 = 260
M1: 128.16
SS1: 104135.99
s21 = SS1/(N - 1) = 104135.99/(261-1) = 400.52
One measurement of the smallest diameter which touches the outer edge of three of the four CRD.
N2: 131
df2 = N - 1 = 131 - 1 = 130
M2: 122.42
SS2: 47925.59
s22 = SS2/(N - 1) = 47925.59/(131-1) = 368.66
T-value Calculation
s2p = ((df1/(df1 + df2)) * s21) + ((df2/(df2 + df2)) * s22) = ((260/390) * 400.52) + ((130/390) * 368.66) = 389.9
s2M1 = s2p/N1 = 389.9/261 = 1.49
s2M2 = s2p/N2 = 389.9/131 = 2.98
t = (M1 - M2)/√(s2M1 + s2M2) = 5.74/√4.47 = 2.71
The 2-tailed t-value is 2.71485. The p-value is .006925. The result is significant at p < .05
Negative stained SP-D dodecamers and multimers: size
I have measured the size of the SP-D images in the article by Perino et al, and their assessment of whether SP-D assists in controlling infection with vaccinia virus. The purpose was to assess whether there was a difference in the size of the molecules from one figure to the next, (which there ws not : Fig 4c-molecules 1-2 =118.71 nm +/ 7.8 nm; Figure 4b 1-11 =118.73 nm +/ 8.76 nm; Figure 4b2 molecules 1-9; =117.27 nm +/ 5.94 nm; Figure 4b3 molecules 1-4; 115.52 nm +/ 4.74 nm. They provided bar markers for each of the images and this was used to determine the arm lengths and total diameter. (SEE MY figure below).
This is the first negative stained group of SP-D dodecamers and lower number multimers that I have measured using the line-node method and the diameter method. While the text appears to suggest that this is recombinant human SP-D (just like Arroyo et al says their dodecamers are recombinant human SP-D) the difference in size between the AFM (latter paper) and negative staining (Perrino et al is huge) i.e. 30 nm approximately.
Figure below shows exactly what molecules looked like, and exactly where i anticipated the molecules to begin and end.