Peak counts of a single surfactant protein D molecule (an AFM image of a dodecamer): Peaks counted in trimers – all processing methods are summarized here. The mean peaks per trimer can be an odd interger depending upon whether the N termini peak is actually one peak, or two, in which case the number of peaks per trimer must be an even number. Both variations occur, and it begs the question of how the four trimers are linked in the middle (ie the 12 monomers at the N termini junction)… some say it varies, and i think i would have to agree.
Variation would be a partial explanation for differences in the center area of the N term peak group in terms of small peaks at the very high point of the N term.
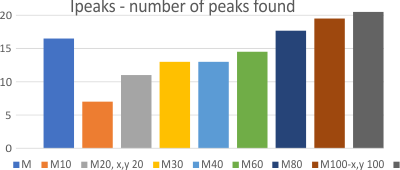
That is a separate topic, this is just total number of peaks per trimer. I used the same molecules, and color scheme, and dataset that has been used in previous posts concerning peaks, per surfactant protein D molecule (trimers, hexamers and dodecamers). Programs used for image processing are listed on the left. Signal processing can be found in previous posts, and i also included the citizen scientist peak counts.

The difference in positioning, and stretching molecules (right and left, or top and bottom) makes a difference in how many peaks are found. This generally is completely known to the observer, the differences in trimer length can be easily seen (as stretching, folding, or compression) and documented. In this particular case the left half of the dodecamer image (arms 1a and 2a, have fewer peaks counted by all peak counting methods) and the right hand side of the dodecamer has more (please look up this link to see the image) (and when i calculate the nm length it will be obvious there too). There are a couple comments on why this happens,
1) is that the extra spread shows the lumpy nature of the glycosylation peak and the adjacent slightly smaller peak.
2) the CRD and neck peaks tend to be exaggerated because of the many ways that part of the molecule can fall onto the mica, and
3) the N termini junction of the dodecamer often then shows a peak right at the high point of that N term peak and the
4) tiny peak in the valley either side of the N termini peak, is often overlooked because of the overwhelming width and height of the N termini peak. Aside from being concealed by the brightness (high gray scale value) of the N term peak, signal processing is very inefficient in detecting tiny peaks when either side has large peaks. So machine learning may be the only way that this tiny peak can be varified. Peak value for the right hand arms of the dodecamer is almost 9, while the left hand is just over 7.