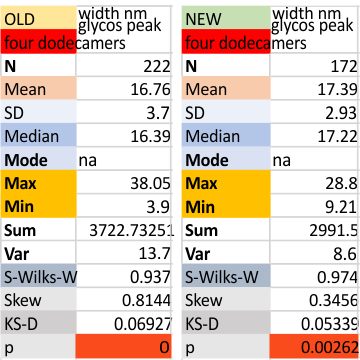
Two different datasets (gathered with the same process, but just different plots) look so similar that it seems likely that this 16nm value for the width of the glycosylation peak will be appropriate for the training plots to examine the other 80 dodecamers of SP-D for which i have pictures.  The valus for all plots together (not divided into the four dodecamers) for old and new datasets below. Please note that the red box at the bottom is an indication of deviation from a normal distribution of the dataset, this shows a significant skew which appears because of the variations in the way signal processing programs divide up the plot into peaks, and how I arrange the peaks into a number (15) that was determined by those same image and signal processing algorithms.
The valus for all plots together (not divided into the four dodecamers) for old and new datasets below. Please note that the red box at the bottom is an indication of deviation from a normal distribution of the dataset, this shows a significant skew which appears because of the variations in the way signal processing programs divide up the plot into peaks, and how I arrange the peaks into a number (15) that was determined by those same image and signal processing algorithms.
Daily Archives: September 26, 2022
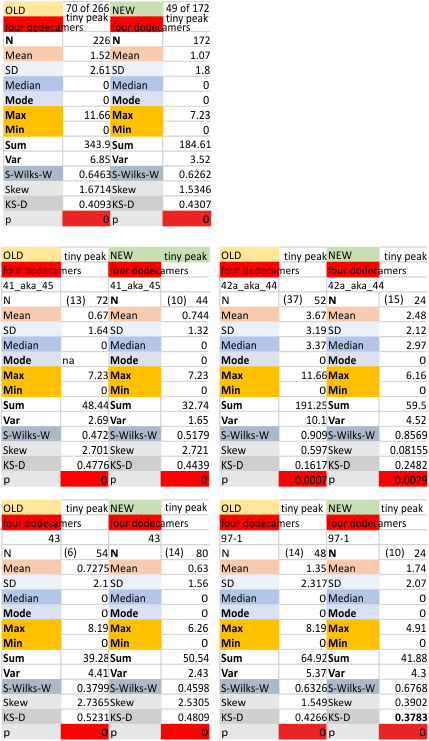
Four Dodecamers: tiny peak (peak 2, lateral to the N termini junction peak) of SP-D
Here are additional, and hopefully the last, set of numbers for Peak 2 (tiny peak beside the N term peak) of SP-D, in terms of peak width. I have used all numbers, including those which do not detect the peak, generated by plots of the trimers (as part of a plot for a hexamer, one CRD to the other CRD to determine that particular peak width. I personally think that adding the missing values is a bit nuts but here are the data, including a new set of numbers which indicates that the peak is a non normal distribution.
It is fair to say that the peak width (when measured only on the values present (please note that the number of actual counted peaks is in parenthesis by N) is N=4, x=3.75+/ 0.34nm, which is consistent with measurements from the beginning of this study. For one dodecamer (42a_aka_44 see below on right hand side) this tiny peak was detected in more than half of the trimers plotted).