Values with and without the “0” missing peaks are given for all peak plots, and for individual dodecamers as well. To me it makes no sense to show the values with undetectable peaks, but it is here (along with the number of times the peaks dont show.


All posts by thankusc
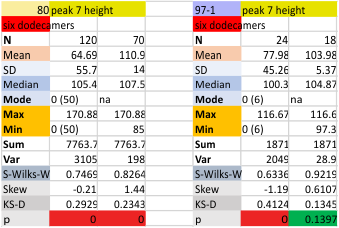
Six dodecamers: SP-D – peak height (peak 6)
Six dodecamers: SP-D – peak height (peak 5)
Peak five data:




data from four peaks here.
Six dodecamers: SP-D – peak height (peak 4)
Only 3 of the 368 total peak values for peak 5 were absent or not detected. I still calculated the numbers with and without the zeros. There was very little difference either way.
Top graphic is the total count, and the mean for the individual dodecamers, n=6. Again, not a big difference, but clearly the n=6 gives the best values for variance and skew. Bottom three graphics are the individual values for each of the six dodecamers for peak 5 height.
Values for the first four decamers is posted here.
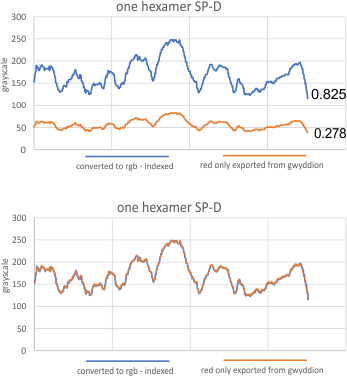
Tiff image from Gwyddion as R, indexed to tiff RGB in imageJ, grayscale plots — same x but different Y
Dilemma: How to use the grayscale plots of SP-D hexamers obtained in ImageJ from images exported from Gwyddion (as red only) in the same datasets as imageJ plots of SP-D hexamers obtained from tiff files exported as RGB. ??? I have left out of my peak height and valley analysis all those plots from Gwyddion because i did not know how to use them. They have very low grayscale peak points and can’t be used along with those which have a highest peak grayscale value (RGB) of around 250. The peaks at about 90 (0-255) and peaks at about 250 for both types of plots just dont work together and I hate to ignore the plots from Gwyddion (as they have a good “limit range and gaussian blur” filters).
To see if I could safely adjust the plots required figuring out, in ImageJ, how to save a segmented line drawn in an image saved in R only, and recall it, and use it on an identical image saved in RGB. So I did this, and while the two plots are not totall “identical” as i moved one node at the right end, they are almost identical. Each grayscale line plotted in imageJ for the R and RGB image was saved to excel. (i wish i could figure out how to create a standard plot template in excel, because even if i choose 0 to 255 scale in ImageJ, excel does what it wants with the y axis and i have to rescale it.)
Below are images of the identical SP-D image (named 127 aka supplement 4A) exported from Gwyddion (gw) as a red tiff, and plotted, and changed to indexd RGB in imageJ, and plotted again using the same segmented plot line restored. Both plots were saved, and opened in excel and a chart was created. Those two plots were saved as metafiles and pasted into corel draw, ungrouped and the line from the R only plot was scaled (without rescaling the x axis) to the same height as the plot from the RGB image and then moved to bottom right of the RGB plot.
As you can there is no difference between the R and RGB plots. So this means to me, that i can take my gw plots and scale them on the y axis and use them in my dataset with the RGB plots. Any issues that i am missing that say “dont do this”?
Top two images are the images with segmented plot lines created (and saved) in imageJ. Bottom image is the two plots, and the lower plot scaled to the y axis (only) and pasted into the RGB plot. — so the difference in grayscale peaks can be scaled using a formula.
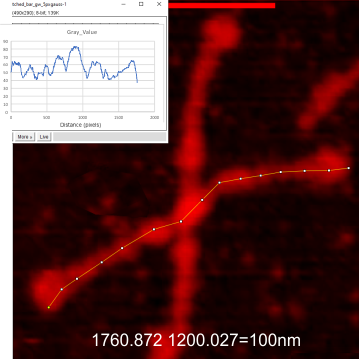
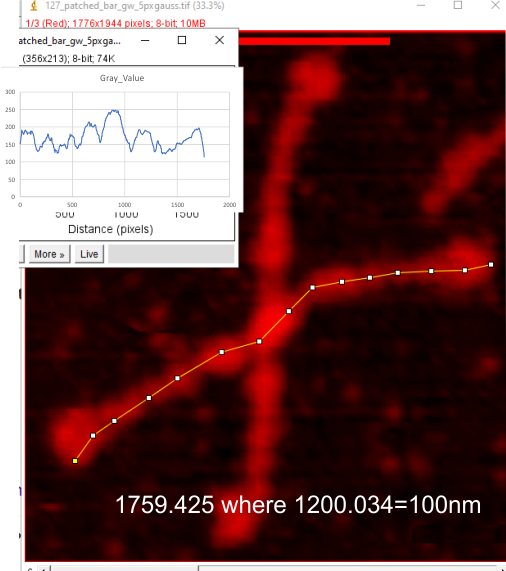
Indeed it would be almost laughable if it just requires a 300pc increase, i would need to find the grayscale value for the highest peak of each image i think inorder to align the R plot to a value.
Thanks to my kids…. dan and aaron. Dan was right in seeing that the grayscale R image max grayscale value was 85. i will use a factor of 3 when analyzing the other gw plots to find a grayscale comperable to the RGB plots.
Six dodecamers: SP-D – glycosylation peak height (peak 3)
Glycosylation peak height. Six dodecamers as individual trimers. N peak is counted full with each trimer. Same MO as many previous posts: y axis is grayscale 0-255, this peak is almost always prominent, one either side of the N term peak and present on most SP-D dodecamers (exceptions being when glycosylation is not present). Check out value from the first four dodecamers, here.
Six dodecamers: SP-D – tiny peak height
Six dodecamers: SP-D – tiny peak height (peak two, in each trimer, located (about half the time) on the down-slope of the N termini junction peak (peak 1) and just before the glycosylation peak (peak 6). This is detected by many of the signal processing programs, at this point it is an unreported peak in SP-D. Means, SD, median, etc listed below for each individual dodecamer of this set of 6, and as a group of 6. MO is the same as reporting on other peak widths-heights-and valleys. First set of 4 dodecamers, 134.7+/19.5 (grayscale 0-255) and all data for that set is here.
Nesting plots trial
Six dodecamers: SP-D – N termini junction peak height
Same procedure, N termini junction peak height for a selected data set (n=368 trimer plots), for the mean of the select 6 dodecamers (all plots each of the dodecamer images of SP-D). The grayscale values of the individual images, does make a difference in the peak heights…. whick at some point i will try to normalize. But for now, the N termini peak has always, and continues to be the highest peak in the plots, typically close to center (there are very rare exceptions which can be attributed to overlapping molecules in an image.
With an extensive number of plots of the first four dodecamers, the grayscale peak was 238.6 +/- 3.75, so the fourth and fifth dodecamers added did change this number slightly. Clearly the values are influenced by the contrast of each image. I think a “limit range” function (which can be found in Gwyddion and likely other programs for image analysis) could be useful. I am more interested in a quick normalizing of peak height and the lowest point in the plot using a graphics program.
(I continue to thank Arroyo et al, and Thomas O’Haver and my two sons, Aaron and Dan for help with image and signal processing apps.
















