Summary of 175 measurements of a single trimer of SP-D (photo taken by Arroyo et al: an image that I call 97): image processing, signal processing:
The aim was/is to determine whether image and signal processing, either combined with each other or each alone, can determine the number of brightness peaks along a plot line of an AFM image of any molecule, with any accuracy. The answer is maybe not!….inspite of the attempt to take an unprocessed image (three different captures), making my own assessment of how many brightness peaks there are, as well as counting peaks from the ImageJ plot, and after using many image manipulation programs, as well as signal processing programs for peak detection ( Matlab/Octave, a Z-score peak detection algorithm and xlsx templates), it is clear that it is a “choice”, simply a choice of what one decides the number of peaks should be. That is not what I had hoped for.
Image here shows the three different unprocessed images used (different degrees of pixelation are evident). These three images were then processed using gaussian blur (5px for 97-1, 10px for 97-2, 15px for 97-2 — which gave them more or less equivalent “look” in my own eyes. Each of those blurred images was also processed with a limit range 130-255 in Gwyddion. Those processed images were resampled in ImageJ at 5000px and 50px (the latter, after some plotting, were discarded as they were just not usable (but i needed to check). Bar marker=100nm.
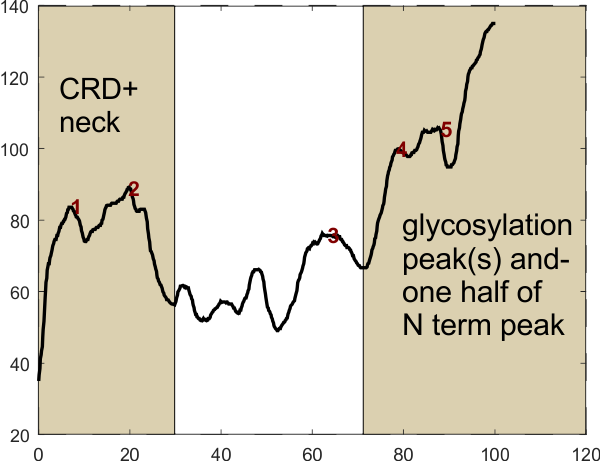
Center image below shows the ImageJ typical trace to obtained the plots, which go from CRD to mid N termini junction. You might be able to see the difference in pixellation in these images as well. It certainly showed up on plots. Plots made in ImageJ were normalized for X, and peak detection parameters for each program is shown by the list at the bottom of this post. 
 The chart above shows every measurement. The number of images plotted is the top row, that is 1-12. The images were number coded so that the investigator did not know which processing was used on which image. Peak counts varied from about 2 to 39.
The chart above shows every measurement. The number of images plotted is the top row, that is 1-12. The images were number coded so that the investigator did not know which processing was used on which image. Peak counts varied from about 2 to 39.
My own determination(s) were somewhere between 8 and 9 peaks, so clearly the parameters for detecting peaks by some algorithms I used was too sensitive. I decided to use the highest and lowest values for what I personally determined the peak numbe r to be (which ranged from 6 to 11) and remove all values from all signal processing results below and above that cutoff. (chart above on right is the result). Peak detection values (from chart on right above) were summed (green insert at left) and the result of the many peak detections (outside of my own estimates) come together in something that is very close to what I had hoped. Yes, i introduced bias by selecting the initial range (someone out there knows a way to do this in an unbiased manner I am sure).
r to be (which ranged from 6 to 11) and remove all values from all signal processing results below and above that cutoff. (chart above on right is the result). Peak detection values (from chart on right above) were summed (green insert at left) and the result of the many peak detections (outside of my own estimates) come together in something that is very close to what I had hoped. Yes, i introduced bias by selecting the initial range (someone out there knows a way to do this in an unbiased manner I am sure).
If you are at all curious about the color coding for the charts above, here is a crib sheet. The greatest number of eliminations came from some values in the Z-LagThresholdInfluence algorithm (8 settings used, some of which sould be eliminated), and the PeakValleyDetection xlsx program where every setting from 1 to 11 was tried (and many could be e liminated there). The Matlab/Octave functions (which I did not write, and have basically no clue about – but listed the specifics below) did not produce as many outliers as other algorithms. Bottom line…. my eyes are best. LOL, sorry, thats just the way it came out.
liminated there). The Matlab/Octave functions (which I did not write, and have basically no clue about – but listed the specifics below) did not produce as many outliers as other algorithms. Bottom line…. my eyes are best. LOL, sorry, thats just the way it came out.
I thank my son, AEMiller, for the batch processing program for the Z-LagThresholdInfluence program.
Each original, and and each image-processed image was also processed by each signal processing algorithm below.
97-arm 1a
my count of peak number from the image
my count of peak number from ImageJ plot
LagThresholdInfluence 1, 0.5 0.025
LagThresholdInfluence 1, 0.7 0.05
LagThresholdInfluence 1, 0.1 0.01
LagThresholdInfluence 5 0.5 0.5
LagThresholdInfluence 5 0.5 0.05
LagThresholdInfluence 10, 0.1 1
LagThresholdInfluence 10 0.5 0.5
LagThresholdInfluence 10 0.5 1
ImageJ find maxima 0.5
ImageJ find maxima 1
ImageJ find maxima 2
PeakValleyDetection xlsx smooth width 1
PeakValleyDetection xlsx smooth width 3
PeakValleyDetection xlsx smooth width 5
PeakValleyDetection xlsx smooth width 7
PeakValleyDetection xlsx smooth width 9
PeakValleyDetection xlsx smooth width 11
PeakDetection xlsx amp t 0.6 slope t 2.5- M6=-4,N6=-3
PeakDetection xlsx amp t 1 slope t 1 -M6=-3,N6=-4:
PeakDetection xlsx amp t 0.6 slope t 2.5 -M6=-3,N6=-4
findpeaksplot x,y,0.0001,80,9,16,3
findpeaksplot x,y,0.0003,0,11,21,4
findpeaksplot x,y,0.0008,40,9,16,3
findpeaksplot x,y,0.0003,0,11,35,4f
findpeaksplot x,y,0.005,5,7,7,3
autofindpeaksplot.m x,y,0.00041228,63.5713,16,16,3
autofindpeaksplot.m x,y,0.00039524,74.8105,17,17,3