I love you children dearly, they were terrific kids, and are wonderful adults. Not perfect, often challenging authority, pushing the envelope, and all the things that accompany being a thinking – motivated adult. This is a treasure valentine from my middle son – just happened upon in the archives of images.
Category Archives: Ultimate order, the cell
This looks simple, but it took a while to figure out… but I can use CorelDRAW for all measurements on SP-D molecules
This looks simple, but it took a while to figure out… but I can use CorelDRAW for all measurements. This program is amazing and I could hardly survive without it. Thanks to my son danny for introducing me to it way back when he was just a kid working at a deli that for some reason used one of the very first versions… CorelDRAW 3. I should buy the upgrade (haha). So measurements of molecules and structures that have been photographed with electron microscopy (or atomic force microscopy, or techniques which shadow or coat molecules to highlight them are rarely made. Some of the really great “oldies” that formulated the basics for morphometry inspired me 40 years ago, and making what is usually a “subjective and only visual” science into something that can be graphed and analyzed has been my goal (forever it seems). So working on the shapes of two surfactant protein molecules (SP-A and SP-D) it became clear that no one really paid much attention to how the represented these molecules graphically… some even represented (and published) them with such “license” (which certainly could NEVER be considered “artistic license” so erroneously that it was adding misinformation to other articles whose authors were using them as models. FAKE NEWS.
It has been great fun working with images of surfactant protein D for TWO reasons: 1) the totally ridiculous diagrams that have been published… i wanted to set straight with something more accurate and 2) these molecules can be very important in the efforts to tailor new synthetic – fullerene-type innate immune molecules to best opsonize viruses and bacteria which enter the lung.
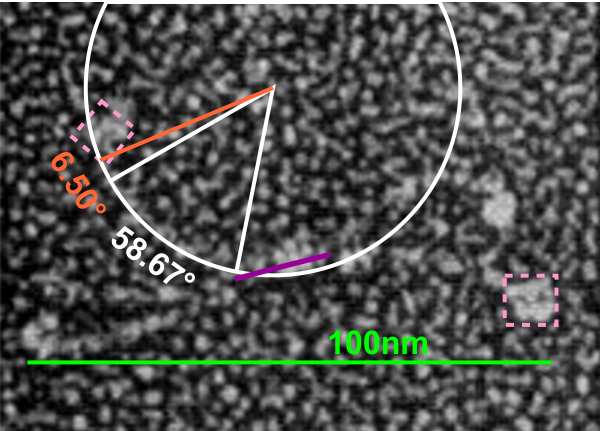
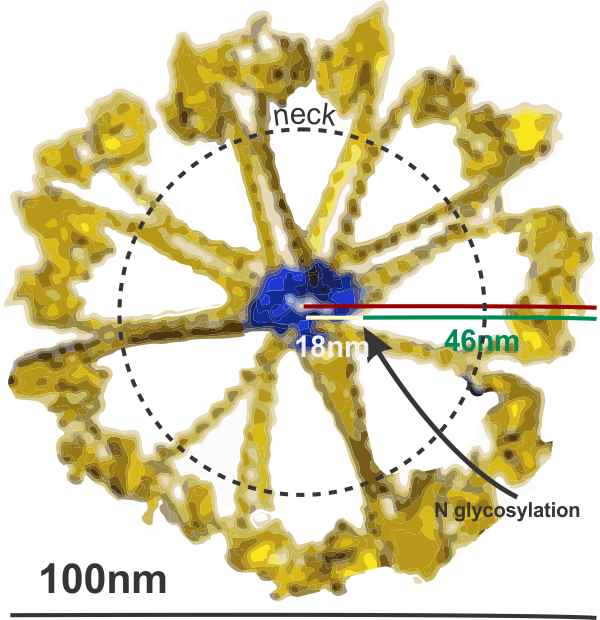
Previous posts show center arc angles in dodecamers, and also the angles of the collagen-like portion of the SP-D dodecamer “arms”, but other things need to be quantified. Diagram below indicates what I think i can measure, and that is the arm length of the collagen-like portion, maybe also the length of the coiled coil neck region, the varying shape-size of the CRD, and the mean length of the N terminal. PER THE IMAGE below.
Using the center arc angle of the collagen-like portion the formula found on this site can be used to determine the arc length. Similarly, the neck arc length. the purple line is from the branch of the dodecamer pairs, and it seems actually to vary quite a bit. THe CRD are easy to measure just as with width and height (since it is a trimeric structure (anticipating from the molecular models on RCSB to NOT have equal width and height, but vary in orientation sufficiently in the micrographs to present with some variation.
There are two kinks for sure, one was predicted by some researchers to be close to the N terminal, and I see one (sometimes represented in various diagrams) at the coiled-coil neck region. I havn’t decided whether to measure that or not. So many studies have shown that the dodecamer is about 100nm that has been used as the standard measure from which other measures are made. EDIT: so neck measures are not going to be possible, and will be part of the CRD measurements.
Verge of a Dream: Like a Serpent
Like a serpent
Wavelessly through
Water without
Apparent attempt
Motionless in cool mud
Dreaming of the
Suddenness of
Repetition
Like an eagle
On a windy rock
Wanting to hang
Like a forgotten picture
Grainy and cool
Without a cushion
Beneath the brink
RLB08151984
Center and arc angles of SP-D molecules
ADDED several new dodecamers to the measure of arc angle of the individual trimeric arms of a dodecamer of SP-D.
N 39
Sum: 1425.97
Mean (Average): 36.563333333333
Standard Error of the Mean (SEx̄): 1.5825376081318
Center angles for surfactant protein D, grouped by degrees (10) do show a tendency to be organized into two sets. You can see that acute and obtuse angles are present. It did require the measurement of more than 30 randomly found TEMs of SP-D to arrive at this graph. Initially no division was seen, the angles looked more like a continuum.
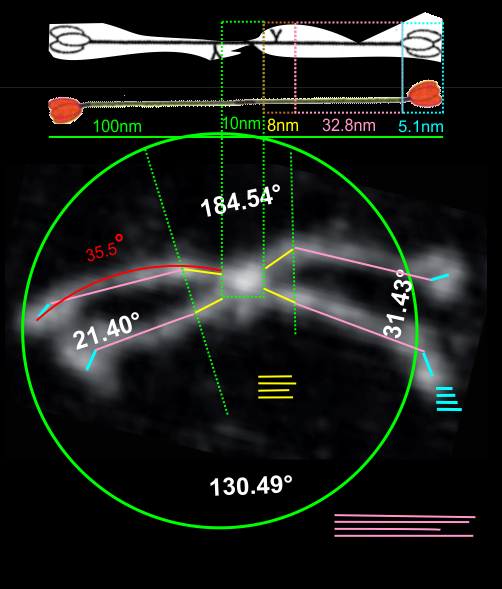
Picture below has several dodecamers added from yesterdays post and it represents several (probably at least 4) different laboratories and at least a few molecules from each. I have created a second “quintessential” SP-D molecule with measures. See bottom figure. It becomes pretty critical to measure the arc angles from the edge of the N +whatever portion of the collagen-like domain remains closely tethered in the center of the SP-D dodecamer. The arc angle changes dramatically with that adjustment. I will measure N terminals of these same molecules (or what everyone describes as the tethered N terminals) and compare with actual TEMs. I can tell you from the diagrams I have found, no one gets that one right when they draw their dodecamers. haha.



Quintessential SP-D dodecamer image
Measuring 30 SP-D dodecamers derived from numerous publications using several different methods for visualization, led me to identify this particular image as the quintessential SP-D dodecamer. The measurements then, from this particular image (most likely from the many images produced by Crouch et al, but which I have also upped the ppi and contrast on) had the closest arc’s angles in each of the individual trimer arms comprising the dodecamer to that which I measured from around 120 trimers. That is…right around 35 degrees. The distance to what looks like a bend and a bright spot in each of the trimeric arms in the electron micrograph likely represent the glycosylation site which was identified by several different investigators. This point gives the trimers the look of an arc. The carbohydrate recognition domains as they attach to the “neck” also like an angle (not yet measured). The center “bright area” of the dodecamer area where all the trimers are bound at the N terminal has a width of about 10nm, at least as is measured in this quintessential image (A collective measure the width and length of the central core of the SP-D dodecamer needs to be done). The the mean for the acute and obtuse angles of the four arms (about 30-50o and 120-140o — in this case the angles are 21.4 and 31.43 and the obtuse angles are 184.54 and 130.49 and so are a not exactly representative but close). 99% confidence level is between 37.00 and – 58.39o for the two acute center angles.
Two commonly reproduced diagrams are included at the top of the real SP-D for comparison. It is obvious that the flexibility of the SP-D dodecamer is clearly missed. And in support of the arms of the dodecamer NOT being stiff, but flexible, authors have suggested that mutant SP-D molecules (shortened collagen-like domains, deleted collagen-like domains, etc) can retain some functions in innate immunity, though sometimes differently than the native SP-D.
The purpose for making the measurements is to determine whether the fuzzyball structures made of dodecamers joined in the center at have the straight arms as depicted in nearly every single SP-D diagram.
Arc angle of trimeric arm(s) of CL-43 compared to SP-D
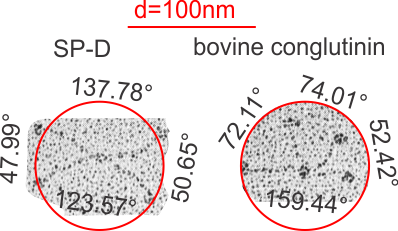
This is a quick test of the arc angle of the trimeric arms of three different collectins but mostly CL-43. The measures are made from publication(s) from other investigators but for this post, principally from Holmskov et al, 1995 so these are not my micrographs. Each molecule (individually) for CL-43 and bovine conglutinin and SP-D seem to display slightly curved trimeric arm(s) (totally unlike the diagrams of SP-D, conglutinin and CL-43. These diagrams are typically straight lines arranged as X s clearly not right. This discrepancy was something that grabbed my interest). Quickly measured arc angle of CL-43 molecules (n= 35) shows an arc angle of 38.3+/1 1.79. This was not significantly different than a couple of dodecamers of SP-D measured similarly (using the best fit circle and arc length to determine the angle) which was n=11 molecules, x=35.7+/- 1.71. I will add the arc angle of conglutinin molecules and a couple of measures for mutant SP-D from papers published in the scientific literature. But here is the single image (replicated 3 times for different measures of the arc angle of CL-43 as shown in the black lines and degree text in each image. The insets are SP-D and conglutinin, with just one molecule each in that inset. Molecules were rotary shadowed according to the authors M&M.
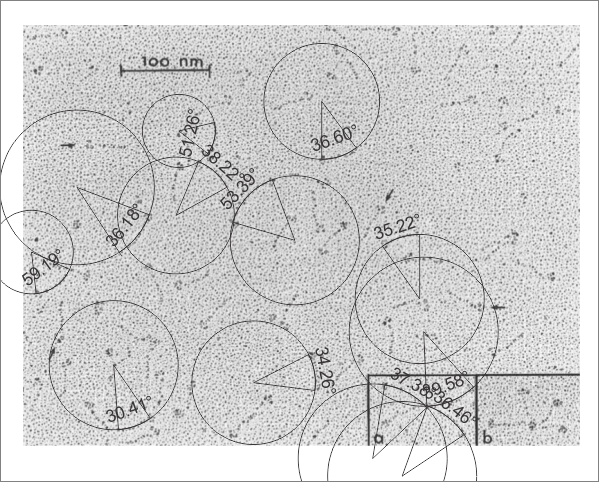
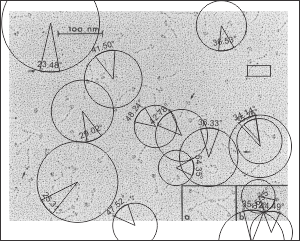
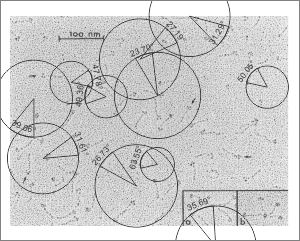 A paired dataset, now with many more samples of SP-D angles than CL-43 shows the two very nearly the same.
A paired dataset, now with many more samples of SP-D angles than CL-43 shows the two very nearly the same.
currently: ONLINE calculator gives these values
Surfactant protein-D
(19 dodecamers – 4 arms measured independently)
N1: 75
df1 = N – 1 = 75 – 1 = 74
M1: 38.11
SS1: 10426.67
s21 = SS1/(N – 1) = 10426.67/(75-1) = 140.9
CL-43
(36 measures = 6 dodecamers – 4 arms measured independently)
N2: 35
df2 = N – 1 = 35 – 1 = 34
M2: 38.32
SS2: 3847.39
s22 = SS2/(N – 1) = 3847.39/(35-1) = 113.16
The results are show no significant difference in the arc angle in the collagen portion between these two molecule types. The next thing would be to compare the aa sequences and aa length of the collagen domain of the arms of these two different collectins. In fact with the current dataset…. the SP-D is right on 38.1 degrees just like CL-43. The calculations us each of the four angles per dodecamer as an N so n= “arc angle”
THE FINAL t-test are as follows:
N1: 35
df1 = N – 1 = 35 – 1 = 34
M1: 38.32
SS1: 3847.39
s21 = SS1/(N – 1) = 3847.39/(35-1) = 113.16
Treatment 2
N2: 129
df2 = N – 1 = 129 – 1 = 128
M2: 35.72
SS2: 20437.31
s22 = SS2/(N – 1) = 20437.31/(129-1) = 159.67
WHETHER calculated with the n of SP-D molecules individually (that is 4 per dodecamer – yes there was one molecule where i could only measure the arc angle of three arms) or whether the t test was done using the mean of the arc angles for the dodecamer as N, where n=30 for SP-D, the results were almost identical.
it will be interesting also to see whether conglutinin is similar…
What to measure, what to compare
In looking at the morphology of a few of the innate immune proteins (collectins) I have been trying to sort out whether the collagen-like domains have a “similar” flexibility, or tendency to “bend”. I was hoping to find some differences, but if they exist I think they will be small (though small does not preclude significance). I found one publication that showed rotary shadowed molecules of CL-43, or SP-D and bovine conglutinin… hopefully at equivalent magnifications (as stated). The micrograph showed trimers of CL-42, dodecamers of SP-D and conglutinin. There is a slight curvature to the collagen-like domain (which is invariably NOT shown in diagrams) which is pretty consistent in micrographs from many publications.
The purpose is to determine if the slight curvature has bearing on how the SP-D multimers associate, and how their trimeric arms are positioned in “fuzzyballs”. The arc angles are shown for each quadrant of SP-D and conglutinin below. THe CL-43 is not shown because it apparently does not form dodecamers.
I am also linking a site for calculating arc angles and other measures of circles as a really helpful teaching website.
Creating an accurate diagram of SP-D —
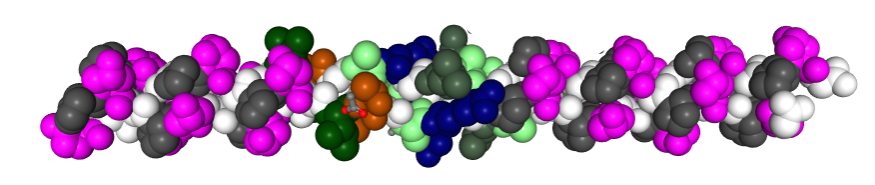
In order to make the correct number of bends in the collagen-like portion of the SP-D molecule diagram which I will use to overlay onto a TEM of one of the many published images of SP-D, i found that correspond to the glycine residues (in this image from RCSB PDB link that has a portion of a collagen molecule that can be viewed in 3D), and the comments on PDB-101, can be used as markers for the angular notations in said diagram.
This is a relief, and i compared the G– sequences in full length collagen (of which there is an easy to count number of at least 360 (that i counted by hand) and a very small number by comparison (29) in the SP-D portion. THus, i will put 29 bands in the diagram of SP-D which you will see is way less than the diagram of Girth Sorensen which I posted yesterday. which indicates there are 171 amino acids in the collagen-like domain. This is problematic, but here is the model of a collagen type triple helix I will use for the diagram.
Some diagrams of SP-D are interesting
I have found that most of the diagrams of surfactant protein D are pretty far from reality. I found this one however that attempts to be more specific and utilizes at least some of the molecular modeling of that protein available. The RCSB PDB really doesn’t model SP-D in any complete way and while researchers have added many many variations of the carbohydrate recognition domain and the coiled coil neck region, nothing else much comes up as to the rest of the molecule. So here is an attempt (which I have vectorized to eliminate the extraneous stuff in the diagram at least it utilizes the structures known for the trimer — from the neck up (haha). Also, it is totally confusing whether the arms are really trimers, or not, because the shape of the CRDs is relatively large compared to the whole molecule and is not tightly intertwined at the neck region (which is the way the “real” molecule is. So Some of these arms are so spread out that there is no intertwining at the neck region whatsoever. So this is not a good representation of the molecular structure, even though it wold suggest that it is a molecular model. So therein lies a dilemma. 1) to make a diagram so wildly wrong that no one thinks you are making a molecularly accurate representation, or do your best, and make mistakes. One thing for certain…mistakes get perpetuated. 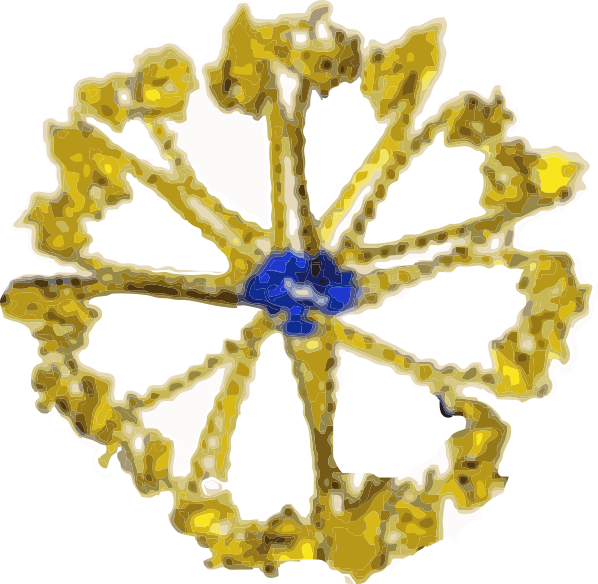
What it does show is the reasonably good approximation of the CRD, and also the curved nature of the SP-D trimeric arms which very few diagrams show but the SP-D trimers and dodecamer molecules seen by TEM show very clearly. What this diagram doesn’t do very well is look at the N terminal portion of SP-D, or show the possibility that there is a connection of the trimers at a “distance” from dead-enter, and it doesn’t indicate where the N terminal glycosylation site might be relative to the whole. Measurements of the latter are taken from a publication by Crouch. Also, it seems that in this particular diagam the collagen-like area here is just a kind of an extended “neck”, with the wrong number of coils, and with no kink, no bend, no separate indication as to where the neck might connect to the collagen-like region. What it also does do, however, is make this particular fuzzyball in a configuration with even numbers of dodecamers (in this case 4, that is 16 arms) which would go along with the TEM images which suggest that the fuzzyballs are an aggregate of dodecamers. What it does not do is look at the possibility of open and closed, or segmented organization of the dodecamers. That would have shown up with more obtuse angles between the four dodecamers and more acute angles within each of the four dodecamer units.
Image found in the following review by Elena N. Atochina-Vasserman
I am suggesting the following structures might have been noted… just by what has been published by Crouch and other researchers on SP-D.
Off-center N terminals of SP-D fuzzyballs
 There is nothing I am doing here but questioning the current notion of how surfactant protein D is arranged into fuzzyballs. I have none of my own images to present, just those published by others which count is close to 75, from several authors.
There is nothing I am doing here but questioning the current notion of how surfactant protein D is arranged into fuzzyballs. I have none of my own images to present, just those published by others which count is close to 75, from several authors.
The publication of Arroyo et al that I have mentioned many times has very lovely AFM micrographs, and they have provided counts of 600+ SP-D images, and it is from these the few of these they published that I am making suggestions that perhaps the N terminals of surfactant protein D molecules are NOT dead center, but arranged in a sort of “ring” near the center instead. The other suggestion is that the arms of the surfactant protein D hexamers (maybe dodecamers) are not straight rods, as almost universally depicted in diagrams of SP-D, but rather are gently curved, almost as if they are hexamers whose interaction at the center is tangent to a center sphere, but at an angle of 60-100 degrees. In measuring the angles in just this one fuzzyball, the average degree was about 67o. (lower left is measured from the dotted lines in upper left — nb, this was just one fuzzyball, and there are many ways to draw those arcs than the way I drew them the first time.)
Image on the left (with its dotted lines) is almost identical to the one posted before (here) but i changed the bending of some of the dotted lines to what I thought was a better fit. In addition I added two more images, with different contrast, and outlined in blue the areas where the N terminal portions of the hexamers make a tangential connection with each other. I think that this (and other fuzzyballs I have observed) pretty much show up as multiples of 2 which is what might be hexamers. Of those which I have currently measured, there will be an angle between SP-D arms, but I will also measure them again, thinking of the molecule itself as an entity which is made up of slightly curved hexamers (or possibly dodecamers), and the angles will certainly be different.
One thing that makes the N terminal attachments NOT being dead center is the presence of slightly brighter areas near the N terminals, which could be representative of the glycosylation sites. For small bright areas to show up at a distance which is proportionate to the ring-center, is encouraging.
Lower right figure has a single hexamer (bent) in which i have measured the angle, and also outlined the bright areas of the N terminal(s), and possibly the glycosylation site(s).
I am impressed by the fact that the CRDs are “lumpy” in this particular image, and I have traced around them, there are almost suggestive of the three CRD domains of the trimer.
One thing that is easy to question on this post is why i drew the arcs with a convex curve and why the sample of a hexamer looks like the curves are concave..haha good question.