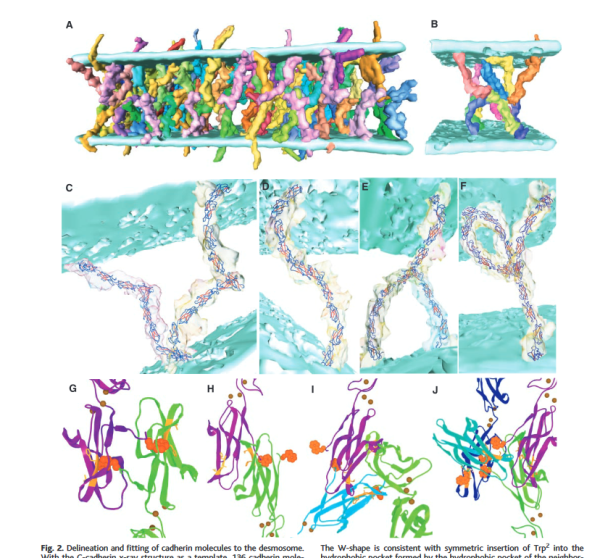
All you guys out there that are doing great research on the molecular structure of the proteins comprising the desmosome, BUT, there is a huge disconnect in what is visible with routine electron microscopy, and the diagrams that are being produced. Just for starters, if I (not being an engineer but just a craftsperson) were going to build a rigid structure binding two adjacent objects i would NOT make it unidirectional. That is I would not orient all the layers of the molecules in parallel. I would do what is commonly done in ply wood, that is to plait the structure with perpendicular, parallel, perpendicular, parallel layers.
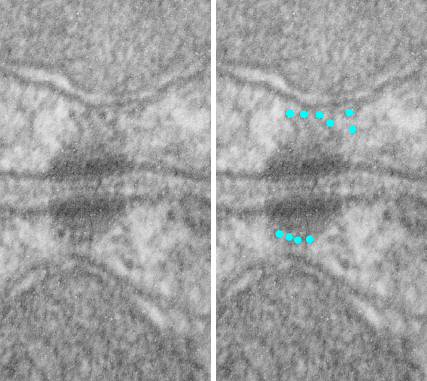
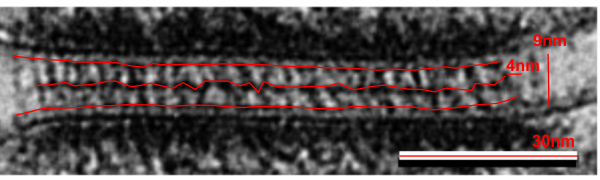
The way i see it in transmission microscopy, the desmosome has a set of proteins perpendicular to (and intersecting) the plasmalemma (desmocollins and desmogleins), then a row of proteins which act like lock washers or lock washers (plakoglobin, plakophilin and others I don’t even know about) that are pretty much perpendicular to the desmocollins and the desmogleins, and then another set of proteins, represented by desmoplakin, that becomes a row perpendicular to the to those “armadillo” proteins, and finally, the connection of desmoplakin with the intermediate (or other) filaments again perpendicular. So there is a layering of different proteins, designed to accept all kinds of stress, and pulling, and shifting.
I have yet to find a diagram, out of literally thousands, which conveys this idea.
What’s more…. the fixation on the desmocollin desmoglein partnership creating the central dense line of the desmosome as being like a W does not fit the transmission microscopy either. It would more likely be an S, again, rarely mentioned, except one pretty close approximation to what is seen (also an S and an Lambda shape).
A really disturbing depiction is the continued showing of intermediate filaments coming into the desmoplakin with a hairpin turn and back out again….. that just is not seen in TEM.
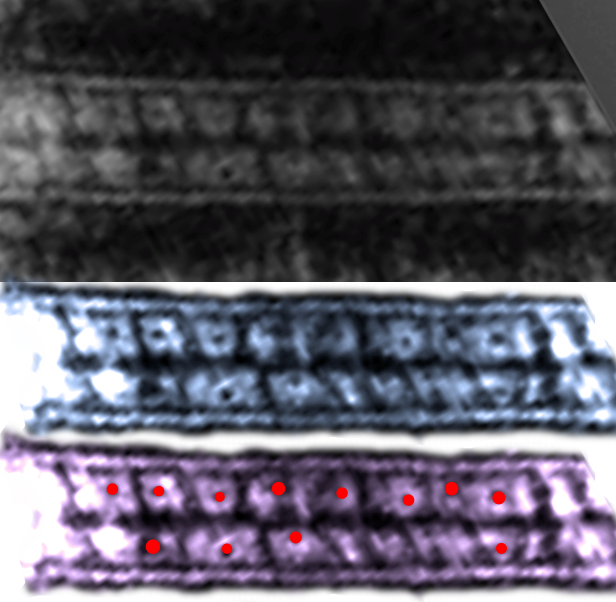
top diagram is what i have readily found in the literature…. mostly parallel depiction of the proteins which comprising the desmosome. (black and colored vertical stripes) — which I think misses the point of the layering. Most left hand bars are what I would predict, everything from the adjacent diagram to the right imply parallel arrangement of the proteins, including the intermediate filaments. 
So this to me is more structurally sound, and we all know nature is pretty good at making changes to improve “soundness” LOL. So I would opt for this interepretation below.  an explanation: plasmalemma (three layers blue and white); central band within the plasmalemma would be what is seen with TEM, i.e. the dense central line (dotted here) made by the association of desmocollins and desmogleins from each adjacent cell and this happens in the extracellular space; the armadillo repeat proteins, as many diagrams put (but not this one) are aligned more or less vertically along with the intracellular domains of desmocollins and desmogleins, but here I have emphasized the “lock washer, or washer” function holding two parts of this cellular rivet in place. THen the desmoplakin molecules which in turn align perpendicularly to the area of the outer dense plaque proteins…. and finally again perpendicular to the desmoplakin… the intermediate filaments (green = direction of orientation.
an explanation: plasmalemma (three layers blue and white); central band within the plasmalemma would be what is seen with TEM, i.e. the dense central line (dotted here) made by the association of desmocollins and desmogleins from each adjacent cell and this happens in the extracellular space; the armadillo repeat proteins, as many diagrams put (but not this one) are aligned more or less vertically along with the intracellular domains of desmocollins and desmogleins, but here I have emphasized the “lock washer, or washer” function holding two parts of this cellular rivet in place. THen the desmoplakin molecules which in turn align perpendicularly to the area of the outer dense plaque proteins…. and finally again perpendicular to the desmoplakin… the intermediate filaments (green = direction of orientation.