This concept of organellar interaction is of course old, and even has been on my mind for decades. I have made many posts on the tethers seen between mitochondria and desmosomes and hope at some point to understand and diagram the relevant collection of outer and inner mitochondrial membrane proteins (black lines above the membranes show area in question) present at the points of attachments to the intermediate filaments (red line shows direction) of desmosomal mitochondrial tethering. Box in top figure is rotated and enlarged for inset. Rat #9, liver 5983, BH
Category Archives: Desmosomal mitochondrial associations
Septate and comb desmosomes – who knew?
Among my favorite papers are those from the journals of microscopy and ultrastructure research way back when the TEM was first available on a commercial scale. Those guys had great eyes, nothing short of eagle vision. The limitations on figures, and words in an article were not so stringent and the micrographs and wordy descriptions abound. This included descriptions of the pretty obvious cell structures and organelles… yep, the desmosome. The potential for examining cells was wide open, and insects, plants and fossils were favorites. The tools were primitive, yes, but the dedication was impressive. I am not too far removed from that group….I would have loved todays tools in last century’s electron microscopy boom. All that “looking” requires time, and in the warp factor 9 world of sequencing and mining….it falls behind. There is, however, no substitute for its value.
So, here is another article from the last century (LOL) showing totally different types of desmosomes (septate and comb desmosomes) the latter of which has layering with a central dense line – a little reminiscent of mammalian desmosomes (in hydra), and also desmosomes (found in silk worm), which actually have some similarities (but big differences) to mammalian desmosomes. Reference is here.
These are amazing structures, however, none of the images shown in this publication have any “look” of a connection with mitochondria as an energy and Ca+ supply, nor do they look like they have connections with the thin intermediate filaments. In fact next to the dense plaque there is a lucent area, then there are the microtubules parallel to the plasmalemma.
One thing that these desmosomes in silkworms have is a central dense intercellular line that was not seen in insect desmosomes in the moth. (vague suggestions of a presence or absence of a central dense line (which in mammalian desmosomes is the interaction of the EC1 domains of two mirrored cadherin molecules that are part of two adjacent cells).
This article calls the picture lower right a hydra septate desmosome. I have to read more of their literature to be sure, certainly they mention the inner part of the trilaminar plasmalemma as being separated more from the outer leafelet, and the outer leafelet being justaposed by dense proteins… (in this case probably analagous to the outer dense plaque in mammalian desmosomes (i.e. the plakophilin and plakoglobin?), and I can see that there is slightly more rigidity to that portion of the plasmalemma. Just for my own edification, i made a mash-up of some portions of one of their figures with measurements made using their own micron bar markers (red bars=50nm). Top image is a “normal” non-desmosomal membrane with membrane to membrane thickness of 18.5nm (brown bar and text) and an intercellular space between the two cells about 13nm (green bar and text). The two images below are their desmosomes. Left lower image looks like a sandwich of desmosomes (three periods=curley bracket) and red bar=50nm which appears to have three iterations (they show pictures of many more than three iterations). The micrograph (from which I have cropped just the desmosome) on the right is single, has two plasmalemma (one from justaposed cells) and a central dense line (see my line drawn in as an overlay). There are differences in the size of the intercellular space of the desmosome and the non-desmosomal plasmalemma above. Pink line is thicker than the non desmosomal juxtaposed cell membranes, extracellular space from outer leaflet of each cell is pretty similar to top image and the central dense line runs about 6 nm in.
Moth desmosomes – nice differences from mammalian desmosomes
I found this paper by Doreen Ashhurst on insect desmosomes which had some transmission electron microscopy of moth desmosomes which are clearly very different than those found in mammals. Brief notes. JCB 46: 421-425, 1970. It is a scanned pdf so the images are not that well preserved but from the text and images here is a set of general similarities and dissimilarities between moth and mammalian desmosomes (typical epithelial cell connections). The list begins at the most intracellular zone to intercellular space, beginning with the cytoplasmic structural proteins, the plaque itself (the outer dense plaque and inner dense plaque in mammals), the plasmalemma, and the central dense line where the cadherins hook up (at least there is a central dense line in mammalian desmosomes, seemingly not in moth desmosomes) .
| WAX MOTH | MAMMALIAN |
| 600nm | 250-300nm diam |
| oval shape | pretty much round |
| cytoskeletal protein=microtubules | cytoskeletal protein=intermediate filaments |
| MT parallel to desmosome | IF parallel to desmosome |
| lucent area by microtubules | desmoplakin molecules by IF |
| fuzzy outer plaque w periodicity | neat tidy outer plaque w periodicity |
| periodicity about 20nm | periodicity about 4nm apart |
| intercellular space about 20nm | intercellular space at desmosome @10nm |
| desmosome has an annulus | desmosome has an annulus |
| intercellular densities periodic | intercellular central dense line |
One thing seems true, the periodicities are less marked in insect (thought the X and Y configurations of what would be equivalent to the desmogleins and desmocollins are just barely seen in this photo by Ashhurst I bet they do have some relationships to mammalian desmosomes. The differning dimensions of the desmosomes between moth and mammal are really quite interesting.
Her micrograph did not have a scale bar but I used a microtubule (with a diameter of 25nm) as the standard. Red line through desmosomal outer plaque used to measure the diameter in the long dimension (presumably) of the insect desmosome. Microtubule in the upper left used to get an estimate of overall magnification of the image (microtubule at a nominal 25nm diameter). Similarly, moth to mammal, there was a slightly reduced width (height) of the intercellular space at the central part of the desmosome, than there was at the extracellular space between the adjacent cells at the points away from the desmosome. The distance within the desmosome being smaller than the distance (height or width of the intercellular space) adjacent but which is NOT part of the desmosome. There is an annulus for the moth desmosome, just like for the mammalian desmosome.
I love that this author says…. “intermediate filaments may run parallel to the plasmalemma in mammalian desmosomes…. ” i know she saw that problem with the early interpretation…. thats fun, and she saw it way back in 1970, and the same is misjudged today. Old habits die hard, and hardly die at all.
Classical and desmosomal cadherins: bullets
Surface proteins
Nidi for cell aggregation
Influence tissue states (layering)
Mediate selective cell-cell adhesion
Regulate embryologic cell sorting
Switches specific cadherin expression
Segregates embryological layers
Image from Farquhar and Palade below, and enlargement
It looks to me like the extracellular domains of the desmosomal cadherins (model found from some other publication obviously) fits well below the plasmalemma of the two apposed cells. This puts the transmembrane and intracellular domains of the cadherins here as a huge portion of the remaining molecule to span from the top and bottom of the punctuated extracellular space (this doesn’t include the dotted central density of the extracellular space generally called EC1) quite a ways from the plasmalemma. I am waiting to find a WHOLE molecular model of desmocollin 2 or desmoglein so i can visually “fit” those molecules THROUGH the plasmalemma on both sides of the cell-cell junction. The red circles, likely the densities at regular intervals of the connection of the intracellular portions of desmocollins/desmogleins with the plakoglobin and plakophilin? There are some very nice tomographic images and studies of desmosomes, but they seem to fit the TEM of the extracellular molecules pretty well (not perfectly) but dont manage to show the transmembrane and intracellular portions of the cadherins with any clarity, in my opinion. And I cant find any molecular images of the whole cadherins (all 3 domains).

More images of desmosomes mostly hepatocytes but many species, many double tethers with mitochondria
The desmosome is not a “spot weld” and not a “bolt” but maybe a modified? “rivet”
A modified rivet
There are many names which have been given to these adhesion points, that is the desmosomes. They occur as spots, yes, of about 200-500nm in diameter, but they are certainly not spot”welds” which would imply that the two cell membranes involved become ONE, ie, the surfaces of the two sides being joined are changed. (This is more reminiscent of the tight junction, matching the spot-weld description somewhat but totally unlike a desmosome).
Others have called the desmosome a BOLT. This isn’t accurate either because the central line in the extracellular space of the desmosme is more like a velcro joint and not a solid inflexible mass but it is a moveable collection of various assemblies of cadherins – mainly desmocollins and desmogleins) where the center portion of the juncture (where the cadherins are bound in a central dense line with a distinct periodicity (something around 6-8nm) can be made and broken depending on ion concentration and cell signaling. This is not at all like a solid bolt-like structure, instead it is more like a zipper. Presumably the bolt analogy which referrers to a something “imagined” but not seen with electron microscopy was just a careless comment since a desmosome penetrating both sides of two adjacent cells is certainly NOT SOLID. A bolt is not only solid but is temporary (or can be temporary) this on the other hand is more like a desmosome.
The desmosome is made up of hundreds of molecules positioned parallel and perpendicular to the plasmamembrane, allowing for flexibility and resistance to various stresses: shear, cleavage, tensile and peel, compressive and torsion. I am sure there are others. The desmosome might be a partial solution to shear, but it would be single shear, since there are only two surfaces involved, one from each adjacent cell.
and ensuring the adhesive molecules penetrate the proximate side of the adjacent plasmalemma of both nearby cells. The bolt analogy doesn’t work since the central joint of the desmosome is easily “made” and “broken” depending upon cell signaling, stage of cell cycle, calcium concentration and sensibility of the cadherin molecules, ie. whether the desmosome is going to be removed or substituted, or unzipped.
- semi-permanent
- cylindrical shank with “head” within each of the two adjacent cells
- similar to a buck at the intercellular junction between the cadherins from each of the two cells.
- major differences is that the “buck” is two sided, like the velcro…. and the integrity of the attachment sis maintained by calcium concentration (and likely many other as yet unknown factors).
A rivet however is defined as a “permanent fastener” so here we have a problem right away…. desmosomes are made and unmade on a regular basis, moved from here and reassembled there to accomodate changes in, activity, stage of the cell cycle, size, forces, maturity of the cells, and countless others. In fact all junctions have to be totally responsive to intra and extracellular events.
Is a desmosome a little like a pop nut, or hollow wall hanger, where the is flexibility in the tethering of cadherins…. just a little, and also in tethering the intermediate filaments by desmoplakin?
Or is the desmosome is more like a bilateral (two headed) unzippable ductile lap rivet.
One of the unique features of attaching two cells with a desmosome that has no parallel in bringing two pieces together in a rivet, is that both sides in the rivet process need to be accessed….which in building….can present a problem. However, intracellular activity makes this access routine and the intercellular components are managed from the inside of each of the respective cells.
I have thought for a while about the arrangement of molecules, and its very nice parallel+perpendicular+parallel+perpendicular line up and felt that this was certainly better than any man-made adhesion rivets…. I wondered whether such a complex set of parts could actually be developed into a real live-functional construction friendly rivet.
Brazing or soldering joint….not applicable in my opinion.
Types of joints….. the desmosome is NOT a butt joint, clearly a lap joint. The spacing of desmosomes just like rivets, will be dependent upon many biological parameters, including states of differentiation.
Great research on desmosomes, but
All you guys out there that are doing great research on the molecular structure of the proteins comprising the desmosome, BUT, there is a huge disconnect in what is visible with routine electron microscopy, and the diagrams that are being produced. Just for starters, if I (not being an engineer but just a craftsperson) were going to build a rigid structure binding two adjacent objects i would NOT make it unidirectional. That is I would not orient all the layers of the molecules in parallel. I would do what is commonly done in ply wood, that is to plait the structure with perpendicular, parallel, perpendicular, parallel layers.
The way i see it in transmission microscopy, the desmosome has a set of proteins perpendicular to (and intersecting) the plasmalemma (desmocollins and desmogleins), then a row of proteins which act like lock washers or lock washers (plakoglobin, plakophilin and others I don’t even know about) that are pretty much perpendicular to the desmocollins and the desmogleins, and then another set of proteins, represented by desmoplakin, that becomes a row perpendicular to the to those “armadillo” proteins, and finally, the connection of desmoplakin with the intermediate (or other) filaments again perpendicular. So there is a layering of different proteins, designed to accept all kinds of stress, and pulling, and shifting.
I have yet to find a diagram, out of literally thousands, which conveys this idea.
What’s more…. the fixation on the desmocollin desmoglein partnership creating the central dense line of the desmosome as being like a W does not fit the transmission microscopy either. It would more likely be an S, again, rarely mentioned, except one pretty close approximation to what is seen (also an S and an Lambda shape).
A really disturbing depiction is the continued showing of intermediate filaments coming into the desmoplakin with a hairpin turn and back out again….. that just is not seen in TEM.
top diagram is what i have readily found in the literature…. mostly parallel depiction of the proteins which comprising the desmosome. (black and colored vertical stripes) — which I think misses the point of the layering. Most left hand bars are what I would predict, everything from the adjacent diagram to the right imply parallel arrangement of the proteins, including the intermediate filaments. 
So this to me is more structurally sound, and we all know nature is pretty good at making changes to improve “soundness” LOL. So I would opt for this interepretation below.  an explanation: plasmalemma (three layers blue and white); central band within the plasmalemma would be what is seen with TEM, i.e. the dense central line (dotted here) made by the association of desmocollins and desmogleins from each adjacent cell and this happens in the extracellular space; the armadillo repeat proteins, as many diagrams put (but not this one) are aligned more or less vertically along with the intracellular domains of desmocollins and desmogleins, but here I have emphasized the “lock washer, or washer” function holding two parts of this cellular rivet in place. THen the desmoplakin molecules which in turn align perpendicularly to the area of the outer dense plaque proteins…. and finally again perpendicular to the desmoplakin… the intermediate filaments (green = direction of orientation.
an explanation: plasmalemma (three layers blue and white); central band within the plasmalemma would be what is seen with TEM, i.e. the dense central line (dotted here) made by the association of desmocollins and desmogleins from each adjacent cell and this happens in the extracellular space; the armadillo repeat proteins, as many diagrams put (but not this one) are aligned more or less vertically along with the intracellular domains of desmocollins and desmogleins, but here I have emphasized the “lock washer, or washer” function holding two parts of this cellular rivet in place. THen the desmoplakin molecules which in turn align perpendicularly to the area of the outer dense plaque proteins…. and finally again perpendicular to the desmoplakin… the intermediate filaments (green = direction of orientation.
Adherens junctions and desmosomes
Narrowing of the intercellular space = about 5nm where the cadherins couple
I am not sure why the intercellular space is recorded by What When How![]() as different dimensions (beside the adherens junctions 25nm vs 20nm, the height of the adherens junction itself) and of the intercellular space beside the cardiac desmosome as 35nm and the height of the desmosomal intercellular space as 20-25nm. It seems to me that if one is comparing intercellular space heights, that one really needs to set some parameters and get some comparisons. This could be a massive job, as there are so many variables, not to mention fixation parameters, tangential sections, membrane proteins, cell types and where on the plasmalemma one is going to attempt to measure.
as different dimensions (beside the adherens junctions 25nm vs 20nm, the height of the adherens junction itself) and of the intercellular space beside the cardiac desmosome as 35nm and the height of the desmosomal intercellular space as 20-25nm. It seems to me that if one is comparing intercellular space heights, that one really needs to set some parameters and get some comparisons. This could be a massive job, as there are so many variables, not to mention fixation parameters, tangential sections, membrane proteins, cell types and where on the plasmalemma one is going to attempt to measure.
Just in four desmosomes (between hepatocytes from syrian hamster) all fixed the same, similar ages and reasonably good section orientation the following is clear.
1. the intercellular space is pretty variable
2. desmosomes are going to have a slightly smaller intercellular dimension than adjacent intercellular space
3. the center dense line extends beyond the outer desomosomal plaque proteins
4. there is a density in the plasmalemmae as an annulus or ring around the desmosome but intercellular space is wide
5. the separation of leaflets of the desomosomal plasmalemma is just a little more distinct than distant plasmalemma
6. the outer leaflet of the plasmalemma at the desomosoe seems to be quite rigid
See below, four examples and the relative wide range of reduction in intercellular space. Syrian hamster — routine electron microscopy, red dots intercellular space remote from the desomosome, blue dots height of intercellular space at the desmosome. Two of the images – right top and bottom – have double mitochondrial tethers. Left side images have a single mitochondrial tether. Right top and bottom have cross and longitudinal sections of intermediate filaments, respectively adjacent to the mitochondria. In both cases the outer mitochondrial membrane also has a rigid look. 
Bridge or intermediate filaments?
When I first looked at this micrograph i thought perhaps the electron density under the mitochondrion (here above two desmosomes, tangentially cut and off center, was a bundle of intermediate filaments. That would be interesting since i don’t know if filament bundles likely composed of keratins 5 and 14, could make this tethering bridge between desmoplakin –part of the desmosome in complex with two remote points on the mitochonrdion.
Turns out that It looks more like a little invagination along part of the plasmalemma of the mitcohondrion, probably representing outer mitochondrial membrane. Red dit=ribosome@27nm, blue curley bracket=bridge, red lines, intermediate filaments bottom red lines, underscoring position of thedesmosomes.
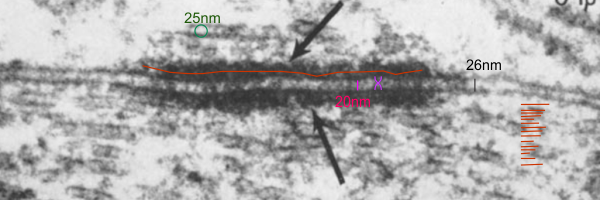
Outer mitochondrial membrane tethered to intermediate filaments
There is a reasonably prominent increase in density (and rigidity) of the inner and outer mitochondrial membranes where they are tethered to intermediate filaments tethered again with desmoplakin of the desmosome complex. It occurs for the stretch which runs parallel to the desmosome and removed from it by about 140nm in height (as measured on these two micrographs – not ideal cross sections of desmosomes, so approximate dimensions).
Someone out there researching mitochondria membrane proteins probably knows which proteins these would be. Top electron micrograph shows two separate mitochondria (tethered — albeit tangentially) to the desomosomal complex showing intermediate filaments parallel to the plane of the plasmalemma in both instances. Red dot, ribosome=@27nm, red lines=intermediate filaments, blue curley brackets=inner mitochondrial membrane with increased density, green curley brackets=outer mitochondrial membrane with increased density at points of tethering. Dotted line in photo bottom left is where the outer mitochondrial membrane likely travels.






