Using a distance and area tool ONLINE to measure inter-ribosomal distance on RER was kind of interesting… There was a freebee trial of 24 hours on this one package called SketchandCalc.com which was in one sense much easier to begin with than ImageJ and L-Measure… neither of which I was able to get data from within the first hour or so. The former unfortunately does not take tif files, or pds so that is a little annoying, but it did import an 8 meg electron micrograph.
It is not intuitive in terms of setting a “denomination” for (for instance) nm, somewhere the measurement got lost? so all measures here are “relative” to my measurement of the diameter of a ribosome (well actually a mean of several ribosomes and I assumed that a mammalian ribosome was 27nm in diameter.
The whole point of making this test was to find the easiest software (shortest learning curve) to make two simple measurements, that is perimeter (and/or distance) and area from my micrographs. I am not happy with the price for the online software since this is a research project and i gain absolutely nothing from it in a personal way, it is not a business venture.
Truth be told i called a local computer repair place (been around for decades) if they might have an old Summagraphics tablet like i used back in the “day” LOL. The owner just laughed.
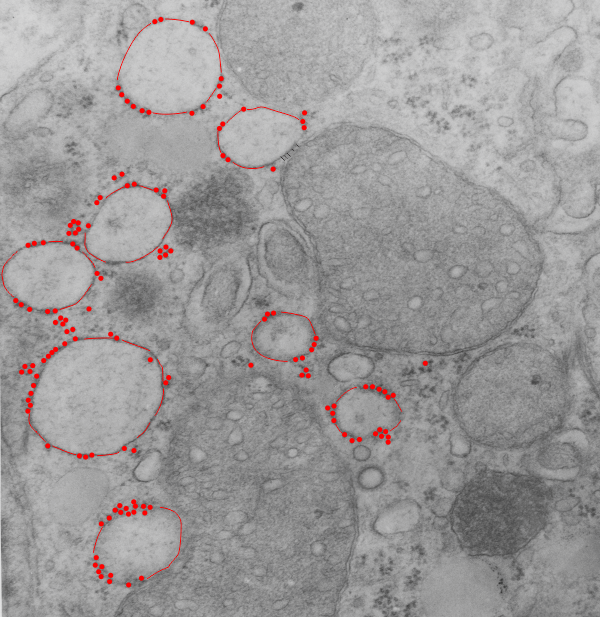
So here is the micrograph, and the measures are of the distance between the attached ribosomes… BETWEEN, because the spacing seemed in to be somewhat regular and I wondered whether there was any biological importance to that. At the same time i took the opportunity to measure several points at a mitochondrial-RER membrane close encounter.
10,000,000 in some cells x couple billion actively synthesizing cells = a whole lot of ribosomes.
Top micrograph (18406_78375_50d_wc-ii no NAC) shows dilated RER (a combo RER-SER with interestingly spaced translocase (?) attachment sites. There are other characteristics too, the length of the string of ribosomes. Red dots are ribosomes (on micrograph below); distance between attached ribosome strings is about 273.6nm +/- 27, n=30, so the SEM is 10%, that is not really that bad for biology and there might be something to this; perimeter of RER vesicles= is about 1500nm, n=10; mean area is about 1600nm2