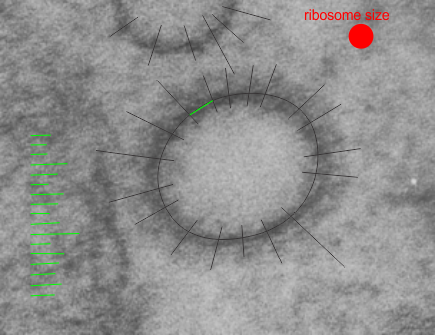
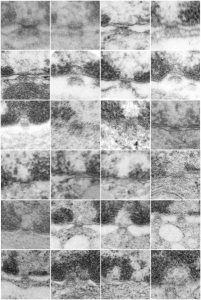
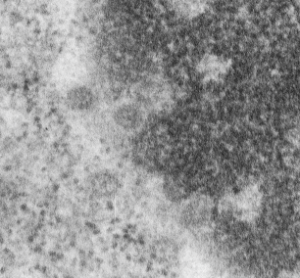
Hepatocyte here, GCLC ko mouse, shows the donut and irregular configuration that is common in circumstances where mitochondria are stressed (I personally have seen it several times but in unrelated experimental circumstances so it is probably a generic response mostly). In this really opportune section one can see a mitochondrion (with an odd donut shape) ont eh left, and a section of RER which has been sectioned tangentially showing the closeness and absolute regularity of the ribosomes along a spiral of mRNA.
I bet at some point all information about these association and the proteins (i saw a list that mentioned in the 800s and counting) in the inner and outer and cristae membranes, and the cristae junctions as well as the matrix will be modeled. Until then, circumstantial evidence for the power of the interactions between mitochondria and other organelles (and cytoskeletal proteins) has to suffice.
The ribosome spirals here look to have approximately 7.3+/- .47 (SEM) ribosomes per spiral, n=9 (a small sample but the best orientation, and a single micrograph…. so this is just a suggested number obviously. 19735_73218_#201 liver alb+/- Gckc -/- postnatal day 28. liver mouse, no NAC.
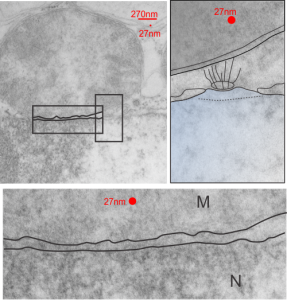
One thing abou these mice that is pneumonic is the dilated ER, a mix of smooth and rough, ribosomes spaced and the presence of the little bubble-blip invaginations of ER within the outer RER membranes. These mitochondria also display fission lines and tubular cristae, and quite a bit of it. Blue dotted line (outline of one part of a mitochondrion – that one tangentially sectioned beside the RER, white box, area for enlargement to the right. Upper image on right has cytoplasmic ribosomes in mRNA-spirals (orange) and lower image is contrast enhanced to highlight the spirals of cytoplasmic ribosomes. I really don’t think there are any good examples of mitoribosomes in either of the two mitochondria shown here.
 Numerous proteins in the outer mitochondrial membrane (encoded by nuclear DNA) target and or are attached to cytoplasmic ribosomes. Cytoplasmic ribosomes have been visualized on mitochondria membranes (that would be the cytoplasmic face of the OMM). These are suggested to be linked by the translocase of the (outer) mitochondrial membrane (TOM) and it is reported that the ribosomes are in clusters (Till Klecker et al, 2014 Trends in Cell Biology) and that pretty much looks like these nRNA-ribosomal spirals… tidily wound.
Numerous proteins in the outer mitochondrial membrane (encoded by nuclear DNA) target and or are attached to cytoplasmic ribosomes. Cytoplasmic ribosomes have been visualized on mitochondria membranes (that would be the cytoplasmic face of the OMM). These are suggested to be linked by the translocase of the (outer) mitochondrial membrane (TOM) and it is reported that the ribosomes are in clusters (Till Klecker et al, 2014 Trends in Cell Biology) and that pretty much looks like these nRNA-ribosomal spirals… tidily wound.
MDVs, mitochondrially derived vesicles (about 70-150nm) are purposeful buds of membrane derived from mitochondria which are targeted to lysosomes, and maybe other organelles.
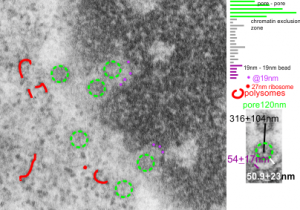
Just an “aside” here, but is it possible that the donut shaped mitochondria (invaginations, and extra turns and indents) might be somehow a deliberate attempt to increase surface area for interaction with other membranes. Another question is the orientation of cristae. I have looked for cristae pores, and that relationship… just not seeing it overtly.