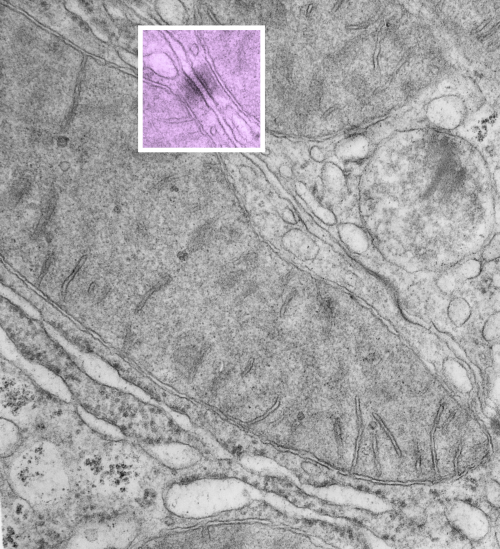
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2211412/pdf/nihms34115.pdfI have marveled at the mitochondria and their propensity to connect with both desmosomes and nuclear pore filaments. These little “bridges” in density and substructure connect them very often. Here is an hepatocyte sample just to show the point. Boxed area has mitochondria in one hepatocyte and mitochondrion in adjacent hepatocyte bridged by a desmosome on both sides. There is great “order to the connection, first a lucent band (disk) then a dense and then a less dense, then a more dense and one more lucent band with little periodicities and a very dense and then lucent band and the plasmalemma. So this is a total of 10-11 gradations in electron density x 2, one on each side of the space between the hepatocytes, and within that space is of course the density pattern therin that is typical for desmosomes. The rigidity of the desmosomal “disk” or “round weld” is quite noticeable, on the plasmalemma side and the mitochondrial membrane side, and the thickness of the whole complex (from the membrane of the mitochondrion in one cell to the membrane in the adjacent cell is pretty fixed as well. I am pretty sure if you search PubMED you will find at least two publications where I mention this phenomenon. I don’t know why it has received so little attention.
This particular mitochondrion also has another connection with a desmosome about half a micron distal to this site.
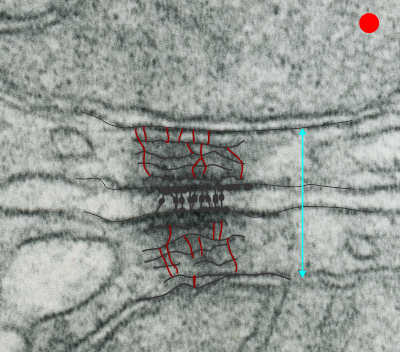
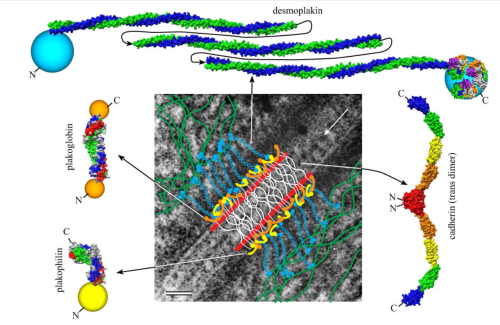
 Making just a line diagram over top of the desmosome-mithcondrial connection i used just lines. Below that is a real great diagram of a desmosome with link to the publication. Ribosome=about 27nm depicted as a red dot. Blue arrow is about 200nm.s
Making just a line diagram over top of the desmosome-mithcondrial connection i used just lines. Below that is a real great diagram of a desmosome with link to the publication. Ribosome=about 27nm depicted as a red dot. Blue arrow is about 200nm.s