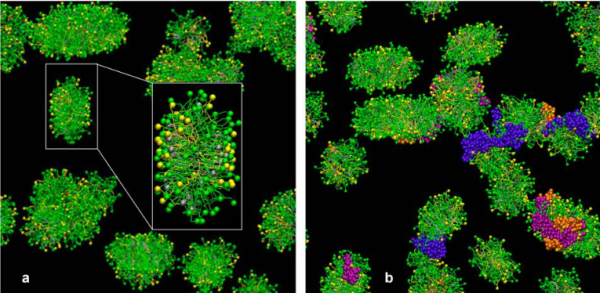
This is kind of a nice article that helps to increase awareness that when we (humans) inhale particles (any particles not just nanoparticles) that we engage those proteins (surfactants initially, both the SPs and the lipids), and the decoration (corona as called in this paper) determines to a degree, where those inhaled particles (nano – man made, or nano natural) will end up… here is their image and a link to the article ., and their legend for this figure: Figure 1. CGMD simulations of pulmonary surfactant (PS) lipids without (a) and with (b) surfactant-associated proteins self-assembled in the bulk aqueous phase. The PS system contains 1400 DPPC (in green), 600 POPG (in yellow), 500 cholesterol (in silver), 4 SP-A fragments (in violet), 10 mini-B (in orange), and 10 SP-C peptides (in purple) in a cubic space of 40×40×40 nm3. For clarity, water and ions are not shown. The lipid contents correspond to a weight concentration of 0.044 mg/mL. Inset in (a) shows the detailed structure of a micelle. What interests me is that they dont mention or show SP-D? and the relative ratios of surfactant SP-A,D and B and C are not reflected in this particular image (i could quantify it with a method of Weibel et al from decades ago). nb, an linked article puts the nm2 of lung surface at something around 100-140nm (not too far off the ones published by the early researchers using TEM to estimate alveolar space.
After skimming through this article i realize it is a but of a summary of meaurements, and a computer simulation.
At least in the second figure (not really reflected in the first) is a size ratio of the three surfactants they chose to model (SP-A (they colored dark purple) , SP-B (they colored orange), SP-C (they colored violet (pink), with SP-A being the larger proportion of the micellar soft corona.
Just wish SP-D had been in their study.
But do you see here an obvious “switch” in the models used for SP-A. In the diagram below they have pictured the whole of the surfactant octadecamer (not just a single trimer) and this model includes all four domains (Nterminus, collagen like, coiled coil neck and carbohydrate recognition domain). It looks on the surface to have the same angular dimensions as the model they give next to it on the right, which is really NOT a good visual correlation and deceiving. Instead the portion of the SP-A molecule they have modeled with their computer algorithms is comprised of just two of the four SP-A domains… thus has nothing to do with the image on the left. I have edited out the coiled coil neck and the CRD from the SP-A octadecamer in the relative magnification that it should be (using their own blue diagram) in the bottom image. The dark purple could actually have been modeled to represent the full SP-A…. it was maybe at least as convincing visual approach.
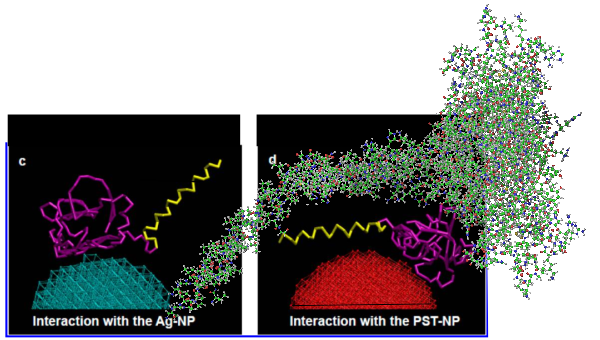
What is missing to me is their identification of the scale… but according to images posted online SP-B is about 60% as big as SP-A (as the octadecamer) but that is a similar length as would be measured by a single trimer (maybe something around 50nm. The polystyrene and silver nanoparticles in this article really are not clearly marked with a bar marker and not easily found in the text…. i have to assume they are something on the order (in relative scale) of 5 to 10 nm which is completely out of scale with SP-A, and would be dwarfed by SP-D (which they did not measure) so this is an enigma.
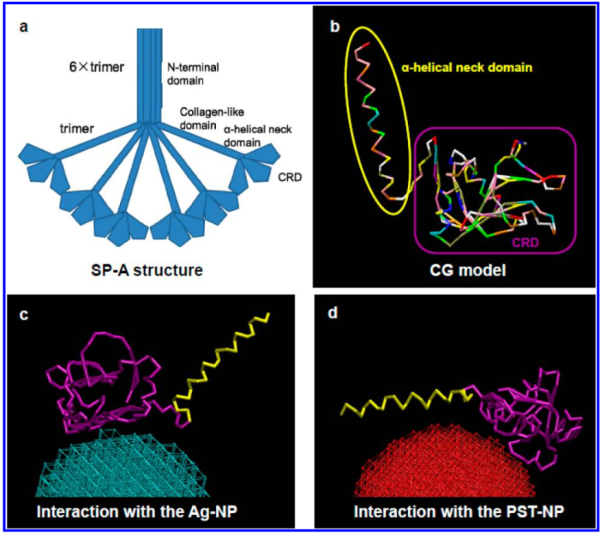
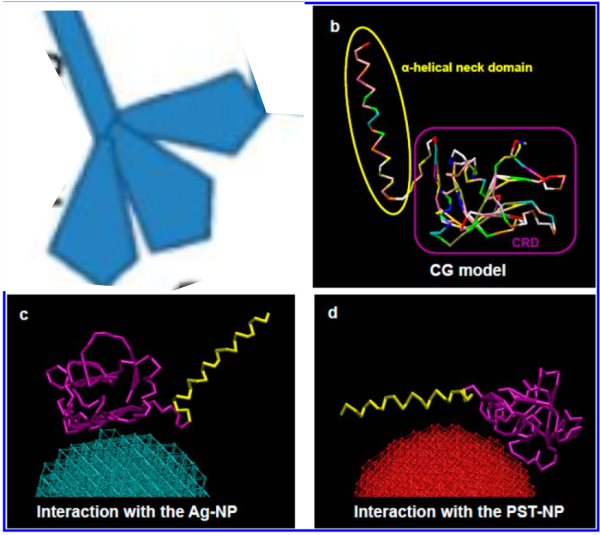 A model of SP-A (which includes an artistic addition of the collagen-like region and N terminus since nothing has been found online that is a model for the whole of SP-A (or SP-D for that matter, and also likely why there is a lot of misrepresentation of both SP-A and SP-D in the literature as to its structure). haha… So if their little red polystyrene nanoparticle is 5nm in diameter, and if a surfactant protein A trimer is 20+nm then the relative ratio of the two would look more like what is below model in green and grey and blue and red superimposed over their image (btw it is the full SP-A, which i diagrammed using the RCSB AND TEM and AFM images posted on line but which has not completely been modeled by RCSB but to me makes sense.
A model of SP-A (which includes an artistic addition of the collagen-like region and N terminus since nothing has been found online that is a model for the whole of SP-A (or SP-D for that matter, and also likely why there is a lot of misrepresentation of both SP-A and SP-D in the literature as to its structure). haha… So if their little red polystyrene nanoparticle is 5nm in diameter, and if a surfactant protein A trimer is 20+nm then the relative ratio of the two would look more like what is below model in green and grey and blue and red superimposed over their image (btw it is the full SP-A, which i diagrammed using the RCSB AND TEM and AFM images posted on line but which has not completely been modeled by RCSB but to me makes sense.