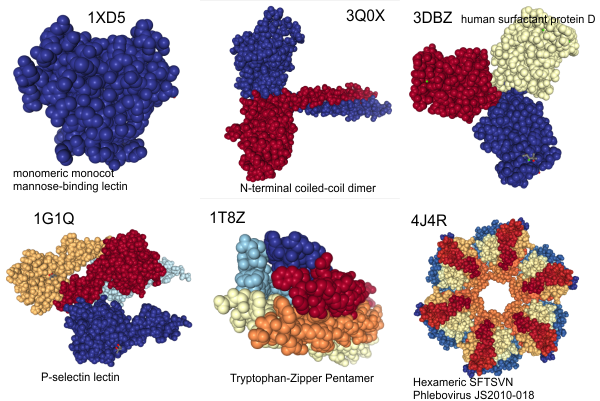
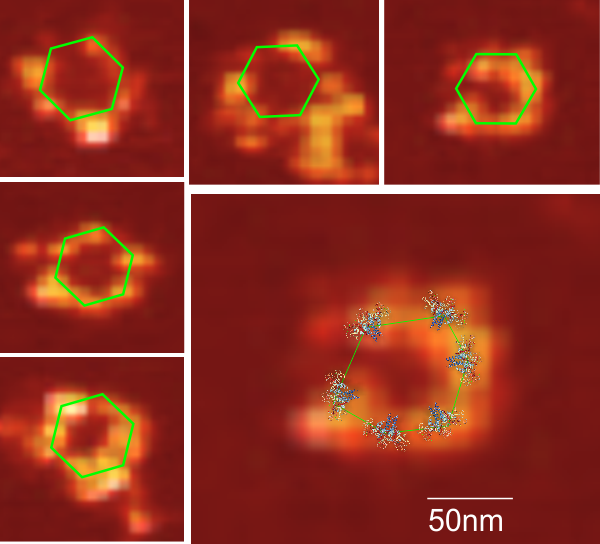
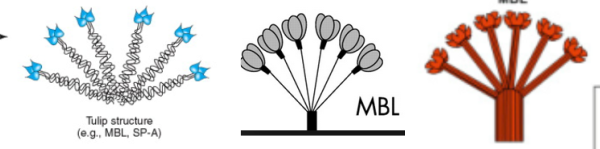
I get kind of sick to my stomach when i read that someone has discovered something that I found (recognized) a decade or more earlier, and know full well that there were people thinking of what i was thinking of even decades before that.Here is a cute article (i call it cute) because it was a worn-out thought in the early 1980s, and I had published at least two papers on the very close connections between mitocondria and cellular elements (desmosomes in particular) and also with nuclear pore filaments in a host of cell types (but mainly hepatocytes)…. Power to this woman, but shame on the editor for not searching back into the 50s and 60s where the wonder of higher resolution electron microscopy was burgeoning to finding out what the real masters thought (and even “moi” was thinking about the connection between organels and possible meaning). This is not to say that the isolation of the proteins and factors was known then, no, but the concepts were there.
What is silly is that I also pseudocolor and isolate organelles to play games with graphically… ha ha. This site is full of examples. oh well.