This is a dilemma which i am thinking could be solved easily, that is: how to adjust the size of the arms of SP-D dodecamers that appear in TEM and AFM images to fit what is understood currently about the protein. The protein is trimeric but occurs, according to publications, most commonly as a dodecamer, that is four trimers linked in the center of the X by the N terminus domains. The measurements of the trimer are given mostly as slightly under 50nm, and the whole can be assumed to be about 100nm (at least this is the consensus number which shows up in publications.
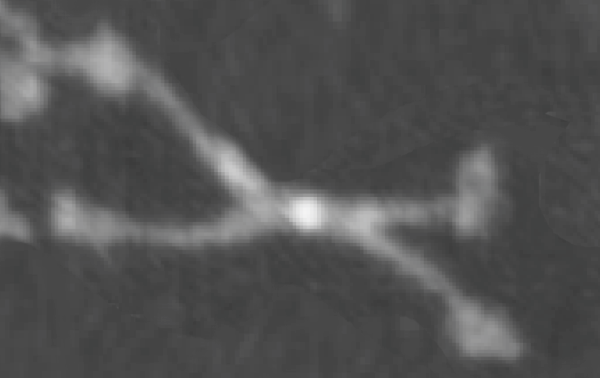
When the SP-D molecules are prepared for microscopy they don’t always fall in optimal positions and this leads to errors when calculating the luminance values since the peak luminance is the N terminus region…and it doesnt always turn up in the very center of the dodecamer (case in point is this published AFM image of a SP-D dodecamer (cant remember whose image it is, but it is not mine – I am calling it image 49 for my own convenience).
two images below are one pair of arms (one complete line of the X – so to speak) cut out from the whole and sliced into 1nm slices and centered horizontally (to allow for use with ImageJ). The top image was NOT adjusted for center (which included delimiting the right and left halves through the N terminus bright spot (orange asterisk) and adjusting each half to 50nm to compensate for the distortion when the molecule was prepared for AFM,) and the bottom image is one which had the two opposing trimers adjusted to 50nm per each using the N terminus (blue asterisk) as center.
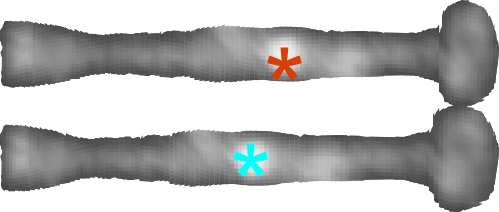
This might appear to biased manipulation of the data, but in fact it is the preparation which presented the bias, and this aims to undo that bias with data that is already known. It certainly helps to uncomplicate the LUT plots.