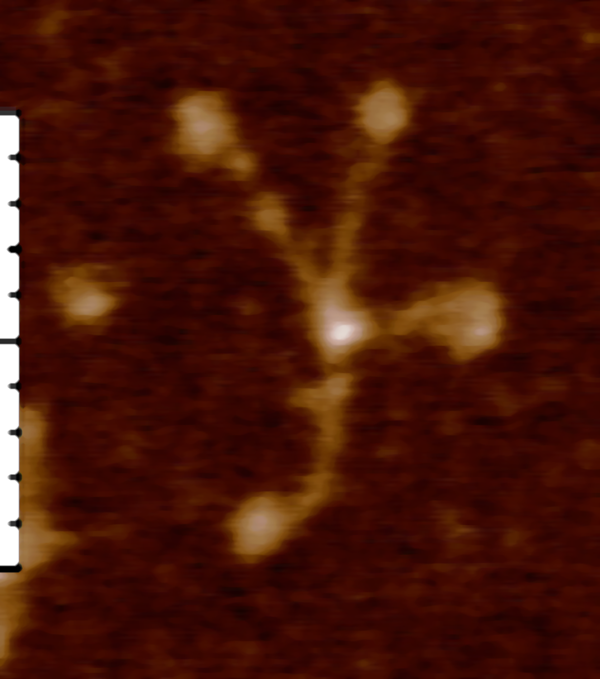
DMBT1 is an interesting multimer and it according to a couple of images I have seen it can appear in two configurations: ring and linear. One confusing issue is why, if the linear molecule has arms apparently extending in two directions along the spine, does the ring multimer not have arms extending toward the center of the ring?
There is a very simple explanation, which I am proposing, not as someone who knows much about DMBT1, but from the perspective of someone who has done lots of imaging, and microscopy and has found that there is an abundance of information in images, it just takes some serious concentration and visualization.
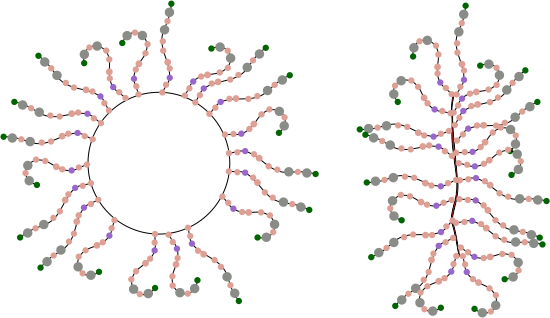
So for starters here are two diagrams (not to scale, not depicting the correct number of SRCR- like domains because I dont really know that number but suspect from looking at AFM images that it is more than 8) and not suggesting that there is a given number of arms in a ring or linear multimer (though the two images I have examined have about 30+ arms each), and I am not suggesting size of the individual proteins modeled nor the amount of space in the link regions (SIR) is accurate, but just making a suggestion about how the ring and linear multimers might actually NOT be so different. At the same time, it eliminates the need for explaining why the linear multimer has arms on both sides and the ring multimer does not.
Below: ring multimer (left) linear multimer (right). If one looks closely at the right image one sees that there is a red fill down the center of the line, which actually I am suggesting represents a collapse of the open circle on the left…. literally stuck together obliterating the space in the ring. This possibility originates from a shadowed image in a publication by Erica Crouch where she attributes the image to John Heuser. The spine of that linear multimer obviously (at least to me) is a junction of two rows.
I suspect that the SRCR domains have the ability to be closely bound, side to side, maybe two or three (judging by the height and width of brightness peaks seen with AFM). If they bind side to side, then why not also bind to molecules across the diameter, more side to side associations, to close the circle. This would look a little more like the sideview of a lampshade with the spine only raised slightly.
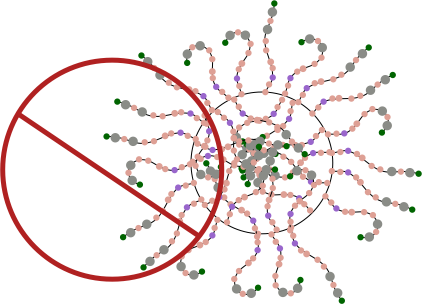
Having the arms on either side of the ring multimer is visually untenable, my guess is that it is an easy leap to suggest that it is not tenable in the molecular sense either. See diagram below. Flattening the ring is about the simplest explanation to explain the side to side arms extending in the linear multimer but not in the ring multimer. See diagram below of the “not really plausible” ring multimer.
For these diagrams, the model for the individual arms was redrawn from a diagram in a publication by Martin P Reichhardt et al, Structures of SALSA/DMBT1 SRCR domains reveal the conserved ligand-binding mechanism of the ancient SRCR fold.http://doi.org/10.26508/lsa.201900502.
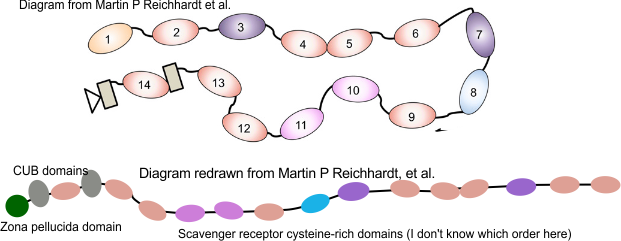

The diagram above (with questionmark) is about the length of each arm that I measured in ImageJ so to make it more or less a reasonable estimate, some of the SRCR domains are not shown (which is consistent with often occurring different numbers of repeats). It seems to me that the linear structure must have some kind of end to end, as well as side to side binding in SRCR domains at the N termini. Just thinking outloud, suggestions are welcome.