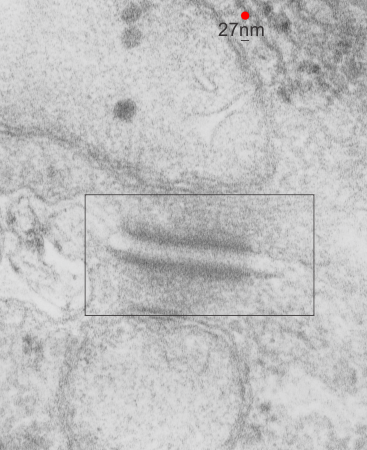
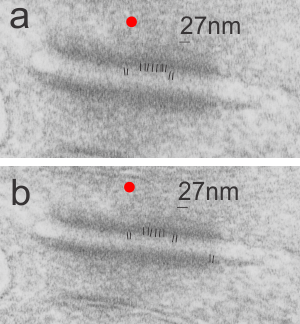
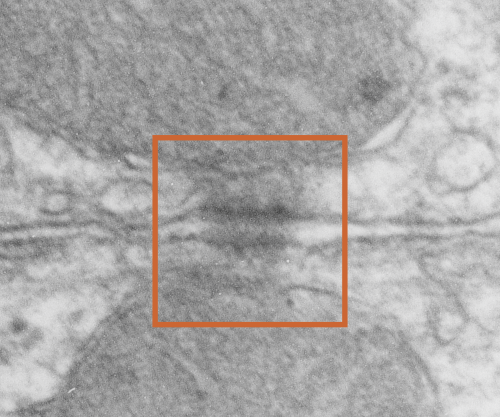
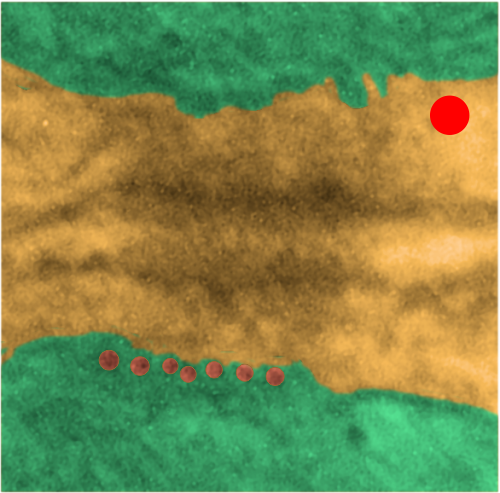
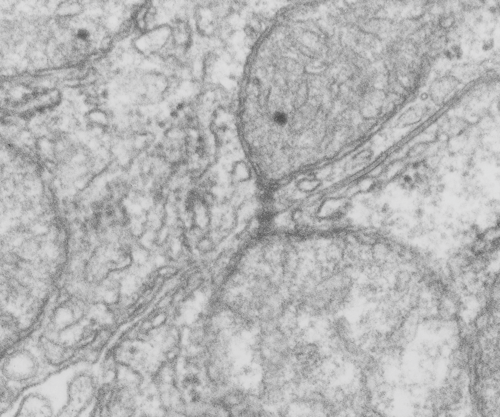
Desmosomes:
(who names these proteins — desmosome.. desmo — and soma, that is the easy button, cadherins… well calcium dependent adhering molecules, i guess thats ok, desmoglobins… not so good, plakophilin, maybe that should be desmoplakoglobofilin.. ha ha… i guess too that the plaque, and plako…. go together, but there are two intracellular zones (apparently) of the desmosome… inner (which is already confusing because to me, if the desomosome is the unit being described, then inner would be the closest plaque to the center of the desmosome, which is the central dense line in the intercellular space, but no…someone named it “backwards” with the center of the CELL rather than the center of the DESMOSOME as the reference point, thus what is named the outer desmosomal plaque is actually “inner” with reference to the desmosome). It is astounding how differently we all describe our world– and it adds a whole lot of confusion and difficulty to learning and understanding).
More or less facts:
1) electron dense disc like plaques (Stedman’s medical dictionary recommends the spelling “disc” for all medical uses) so I will use disc.
2) extracellular-intra-and-intercellular-cell-to-cell adhesion discs
3) prominent in epithelial cells
4) 0.2 to 0.5 microns in the flat dimension (depending likely on whether the edge of the disc is in the plane of section or the center
5) very likely different states of cell activity change the dimension and the number and the location of desmosomes as well as the number of mitochondria which are associated.
6) Only a small percent of mitochondria are associated with desmosomes, mainly because there are many mitochondria in each cell. (Volume vs perimeter consideration).
Desmosomal cadherins – desmocolin (3 isoforms) and desmoglein (4 or more isoforms). DSC2 and DSG2 are the most common.
Tying the desmosome to intercellular elements are the “intermediate filaments: a diverse class of flexible filaments that provide mechanical strength to cells. They make up hair, nail, horn, and scale cells, they form the nuclear lamina, which lies just inside the inner nuclear membrane, they span cells to provide strength to epithelial tissues, and they anchor organelles and stabilize the cytoplasm” with these data (Intermediate filaments 6–10 strands per filament (6 was quoted for those which are next to desmosomes), non-polar, outside diameter he quotes as ~10 nm (I think this might be larger, or maybe stains add dimensions)) an edited quote from Methods for modeling cytoskeletal and DNA filaments by Steven Andrews (Physical Biology, Volume 11, Number 1 )
Desmoglein 2
“Desmoglein-2 is a 122.2 kDa protein composed of 1118 amino acids. Desmoglein-2 is a calcium-binding transmembrane glycoprotein component of desmosomes in vertebrate cells. Currently, four desmoglein subfamily members have been identified and all are members of the cadherin cell adhesion molecule superfamily.”