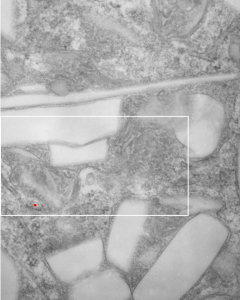
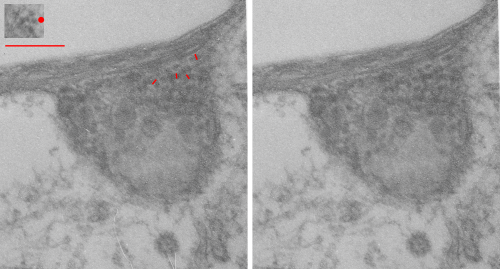
The phagolysosomes in phagocytes which containing perfluorodecyl iodide crystals (in this case the perfluorocarbon has been retained in the body for more than 250 days – mouse liver) show huge amounts of lysosomal enzymes. The collection of macrophages (even multinucleate macrophages) don’t appear to initiate a huge collagen response, and inflammatory cells such as neutrophils, just are not that common, even when phagocytes are plentiful.
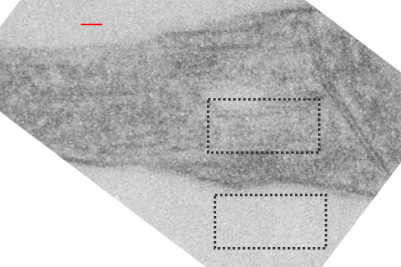
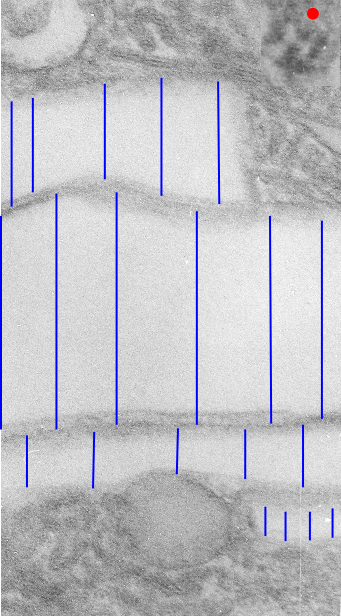
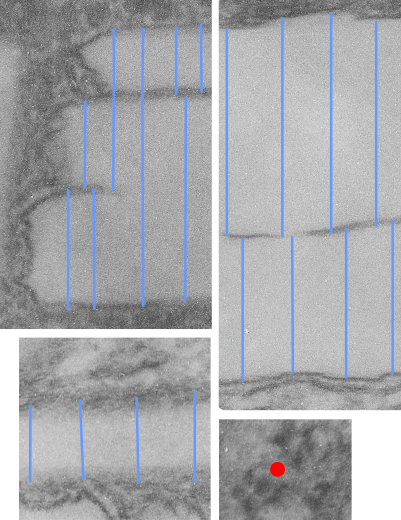
The ultrastructural characteristics of the lysosomal enzymes contents within phagolysosomes containing IPFD crystals can vary dramatically within the same cell and in adjacent lysosomes, and within the same lysosome. This varies from a very fine homogeneous non-textured content in some areas to areas with a more coarse fibril-like texture, appearing like a lumpy (sausage link type) strand or a twisted strand (of two separate elements, or a two strand braid). In some cases the fibrils appear to be solid on cross section, but in other cases more like a tubule. This underscores the variability that would be found in a twisted fibril, allowing for the explanation of both “hollow” and “solid” profiles on cross section.
Transition zones between the types of enzyme ultrastructure are found within the same lysosome suggesting some protein overlap or continuity. There are some areas of very dense accumulation of proteins.
IPFD crystals may be large or small, and most display some obvious directionality, usually the longer side having a thin dense protein (or trilaminar membrane?) coat, but at the ends showing considerable “striping” or dragging out, or whispy periodicity.
IPFD crystals often appear in cross section to be more rounded, or even round.
Some common characteristics of the fibrillar texture of lysosomal contents of IPFD filled lysosomes follows:
- relatively even diameter at widest and narrowest areas
- often looped and tangled, folded looking
- equally even spacing seen as a predominant lucent area surrounding each strand
- often running in parallel
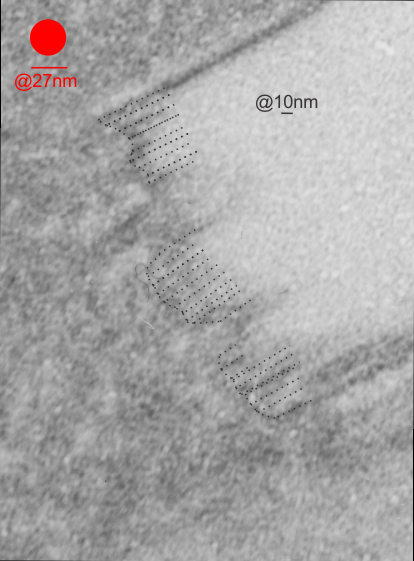
- higher mag shows sub-layering
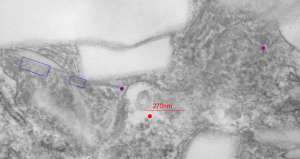
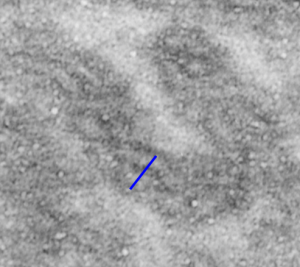
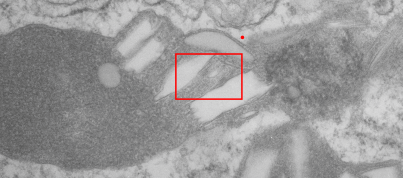
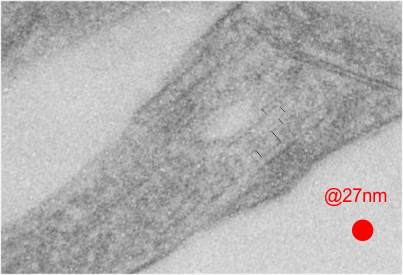
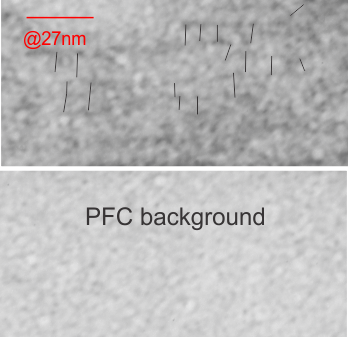
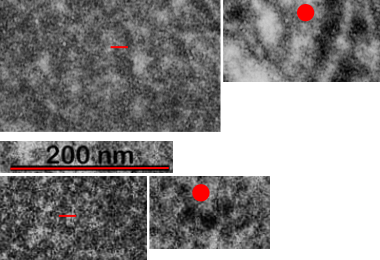
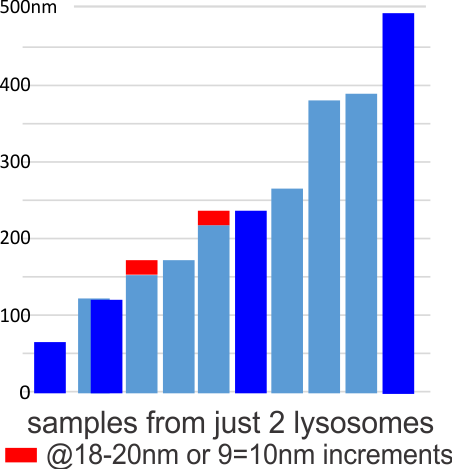
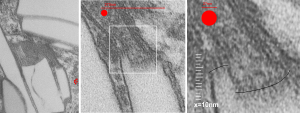
This image below shows a very prominent (yet small in terms of nm) level of periodicity. May not be the smallest since I havn’t be able to find any information on the actual crystal chemical structure of perfluorodecyl iodide), but pretty small for conventional electron microscopy to propose, but this is so obvious that it certainly warrants comment. The single space (inset is same size as lower image and placed on an identical image, enlarged so that the lines (periodicity) could be seen. The span is about 9-10nm line to line, and the fiber arrangement is double that…. between 19-20nm. It appears that the protein(s) oligomerized here can come as an interconnected (maybe twisted) double strand.

Lower magnification image of the portion of a macrophage from which the inset to the right and above (white box) and the measurements of distance between fibrills (white lines)and smallest periodicity (blue box) measurements were made. Red dots=ribosomes at an estimated 27nm for reference; blue line = smallest periodicity; purple line = the 9-10nm pair, twisted, representing the fibrills (about 20nm in thickness if section is longitudinal, or diameter ir fibrill is cross sectioned. A measurement of about 20+nm is found from center of one fibrill, perpendicularly, to the center of the adjacent fibrill, (white connected lines on the bottom micrograph show distances between fibrils measured. Image at bottom has blue line of @20+nm –the approximate diameter of the “fibril”, and the dividing line of the two strands is visible.