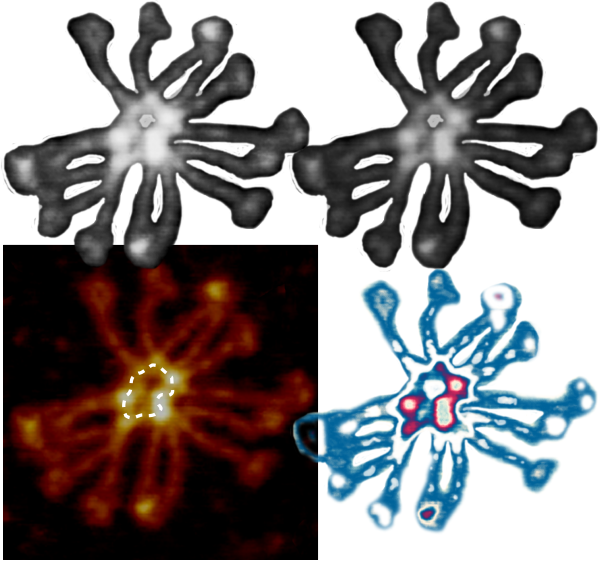
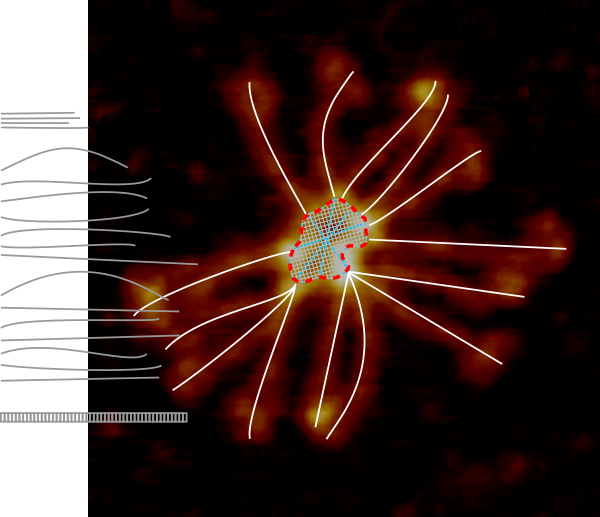
Another post giving the math for converting RGB to greyscale is found here.
It seems that Luminosity, Lightness and Averaging are three methods which are used often. I am looking for someone who can find which of these three methods Photoshop 6 and CorelDRAW x5 uses to convert RGB to greyscale. (CorelDRAW under “autoadjust” and
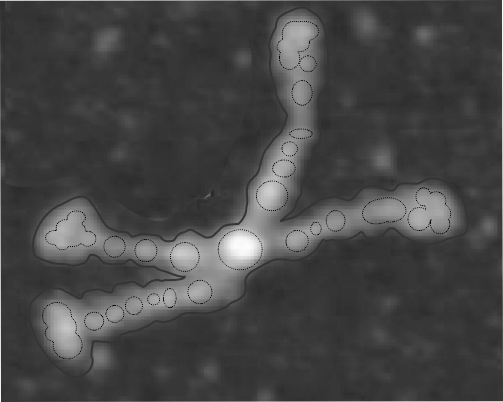
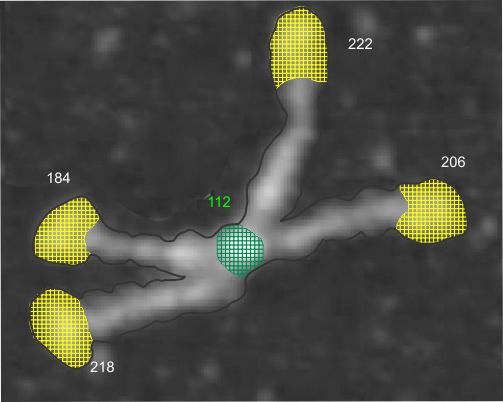
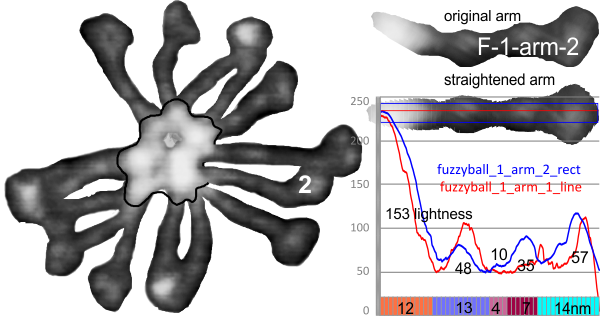
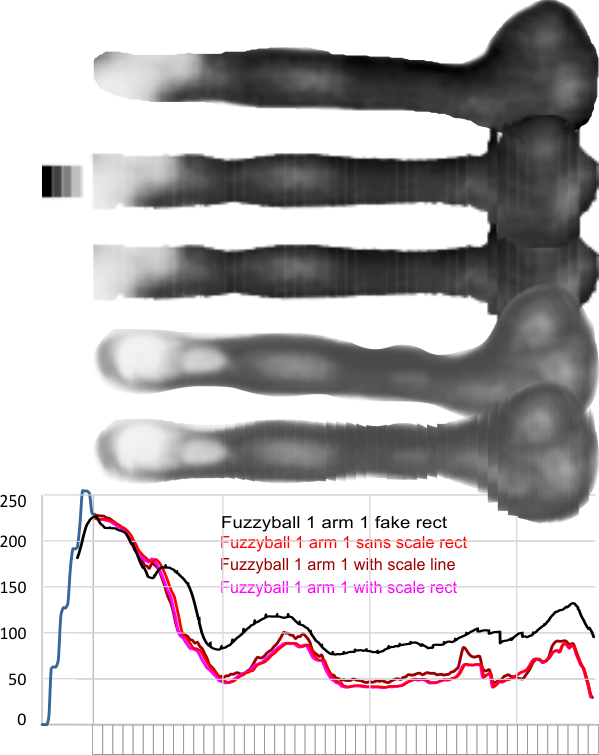
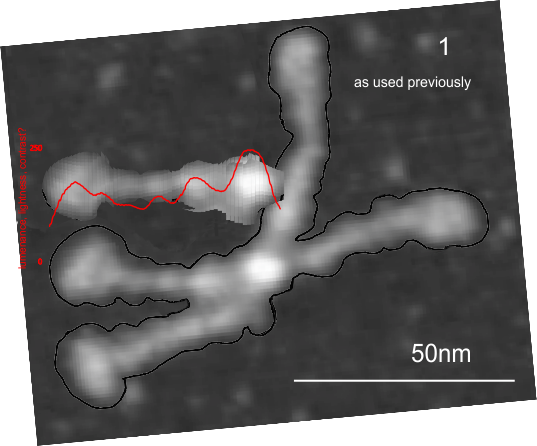
In photoshop 6, thresholding a black and white image just doesn’t really help (in my opinion) because there is too much loss of the greys.Below is an image (used many times before) of SP-D (not my AFM images, someone elses available online) which I have manipulated in photoshop using image/adjust/levels, and image/adjust/autolevels, and image/adjust/autocontrast, and image/adjust/contrast in various ways. I will plot my favorite arm (upper left) which has three peaks between the N terminus center high point and the CRD, having probably appeared more times than any other configuration, and the pic is also biased to my assumption from previous LUT plots, to have three different heights for those three peaks, and also different widths. I will post plots in for that same arm, in the same order below. Top left, image as used previously (test 1) provides a great plot.
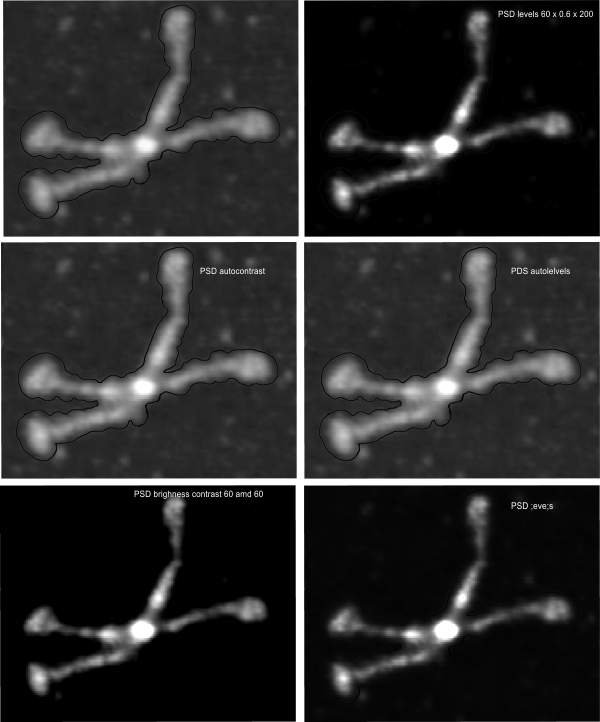
Test 1, image as used previously, not very much adjusted (i cant remember but likely some contrast increased and brightness reduced after the original conversion from color) but done in the same manner as other SP-D dodecamers that have LUT plots (see countless previous posts).
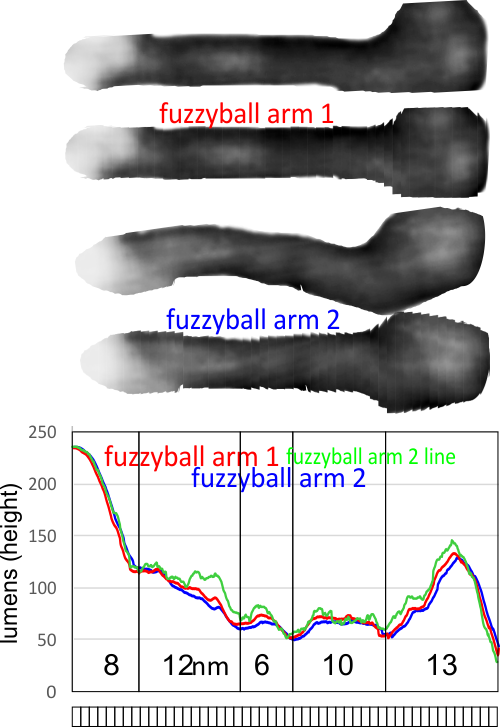
Confirming, and reconfirming previous LUT plots, there is an N terminus peak, three intermediate peaks (in this case the third peak from the N terminus is pretty small) and the bumpy peak of CRD (owing to its distribution of the three CRD domains in a large bulb like end.
Little adjustments, same peak, so that points to the small effect that the several different algorithms for changing RGB to greyscale may not matter as much as I thought. Here is a link to another article on converting color to greyscale. and yet another on terminology and greyscale (ok, so i need to remember to use grayscale not greyscale) conversion.
 While i know to little about this to have an opinion, my basic understanding is that most of the conversion software attempts to mimic what the human ee sees, rather than wht the RGB actually convert to.. in terms of — Gray = (Red + Green + Blue) / 3 — which for science, i.e. looking over micrographs) probably needs to be as free of attempts to increase or decrease the “RGB” values to fit human perception. SO, that said. perhaps less manipulations, fewer algorithms might be the best approach.
While i know to little about this to have an opinion, my basic understanding is that most of the conversion software attempts to mimic what the human ee sees, rather than wht the RGB actually convert to.. in terms of — Gray = (Red + Green + Blue) / 3 — which for science, i.e. looking over micrographs) probably needs to be as free of attempts to increase or decrease the “RGB” values to fit human perception. SO, that said. perhaps less manipulations, fewer algorithms might be the best approach.
I have used RGB conversion in photoshop, in coreldraw, and desaturation to convert RGB to grayscale (yep now gray with an “a”).
After reading (rather stumbling through) some manuscripts and blogs about dozens of RGB to grayscale equations … my decision is to go back to basic grayscale = R+G+B/3 rather than use all the fancy formulas that make the grayscale look more pleasing to the human eye and automatically adjusting the blue and greens to make things look “nice”. Then I can adjust the contrast to what pleases my eye, emphasizes what I am looking for, and brings out the structural detail in my images, manually, and define it as such. To find the codes that were used in some programs is totally impossible, and if found are probably just as biased a conversion effected by my “hand made manipulation” of the images. If the conversion from RGB to grayscale is standard and the adjustments, tweeks are mine I can just comment on them in a line of text in the materials and methods… ha ha
Who knew that was (could be) so complicated. Hahaha…
FOR ME…. my eye is still the best tool to convert and desaturate and change the brightness and contrast. It might very well be biased, but at least it is not brainless.