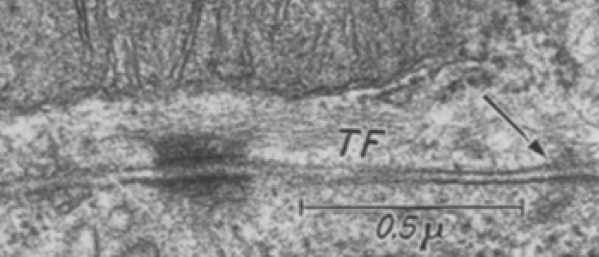
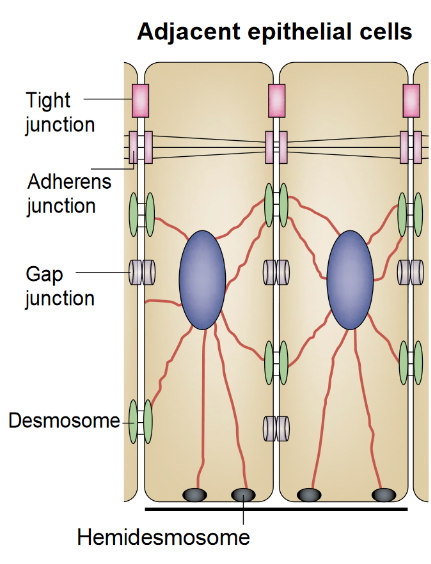
I found a cryo-EM paper of desmosomes that was pretty nice. Well desmosomes in the lower portion of the epidemis might be like desmosomes in liver, and it is very clear from this paper and others that the desmosome is adaptable, become different things in different cell types and tissue types. So this is a general description, but the basics are present in desmosomes of the liver. The measurements they show do NOT exactly fit what is stated for the “viable epidermis desmosome this publication states, but they are pretty close to those that I have re-measured just from their image, their bar marker, and my own repeated measurements from densities to densities.
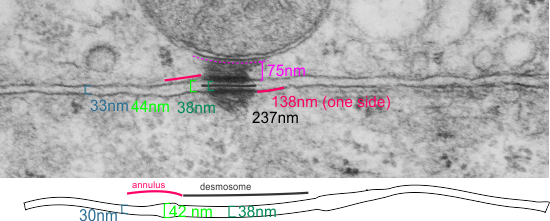
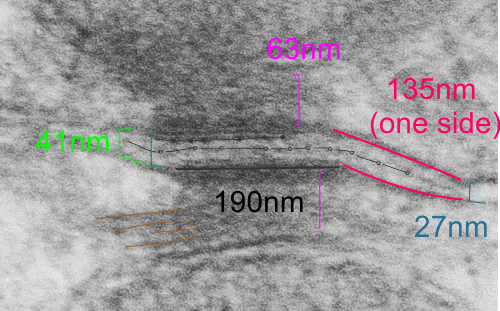
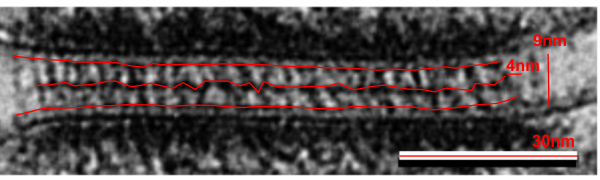
The measurements I came up with are posted on the micrograph. The baseline measure came from the micron marker given for their own image. The measurements I made are for inner lamina to inner lamina of the two plasmalemmas; outer lamina to outer lamina for the two plasmalemmas (extracellular width); the pretty lucent area just extracellularly to each plasmalemma, the length and dimension between the desmoglein and desmocollin molecules (cant tell which is which) and also the “v” type structure, alternating and overlapping as the center dense line of the desmosome (in the extracellular space).
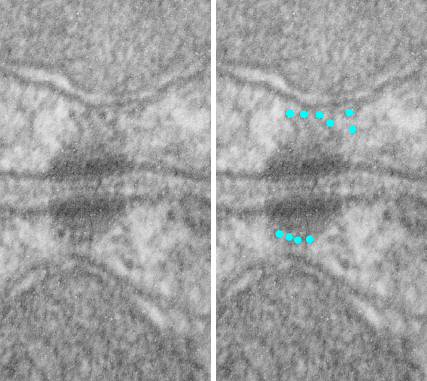
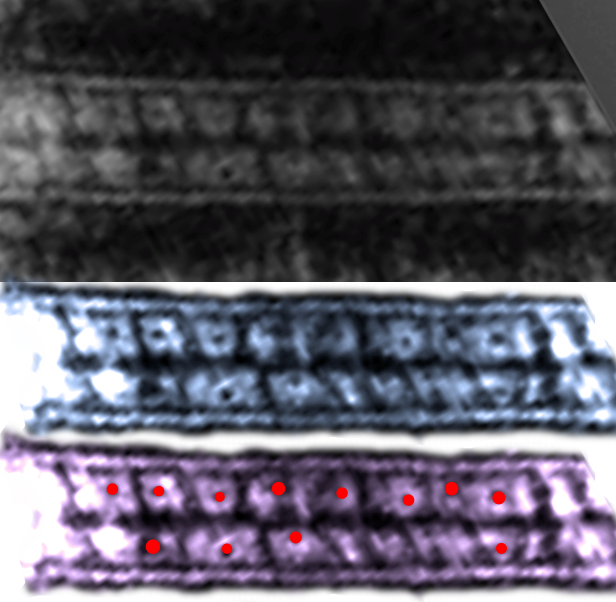
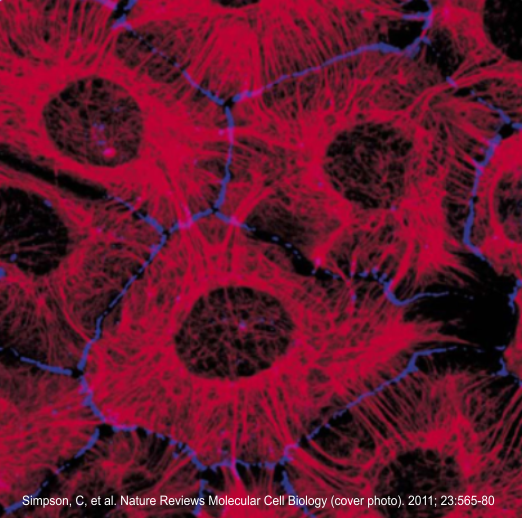
I thought it was interesting that one researcher called this intercellular linear pattern random, while another called the pattern linear…. in fact i think they both missed the pattern and I have outlined the cadherin rhythm in green. The distance between the repeat portions (ectodomains) of the latter looks maybe to be close to linear, or slightly curved, and the N terminals (to my way of thinking) create a picket fence, or zipper kind of pattern.
The micrograph I used for these measurements came from a paper by Ashraf Al-Amoudi, Jacques Dubochet, and Lars Norlen which is available online. Their bar marker is at the top (50nm) and all other measurements were made from this distance.
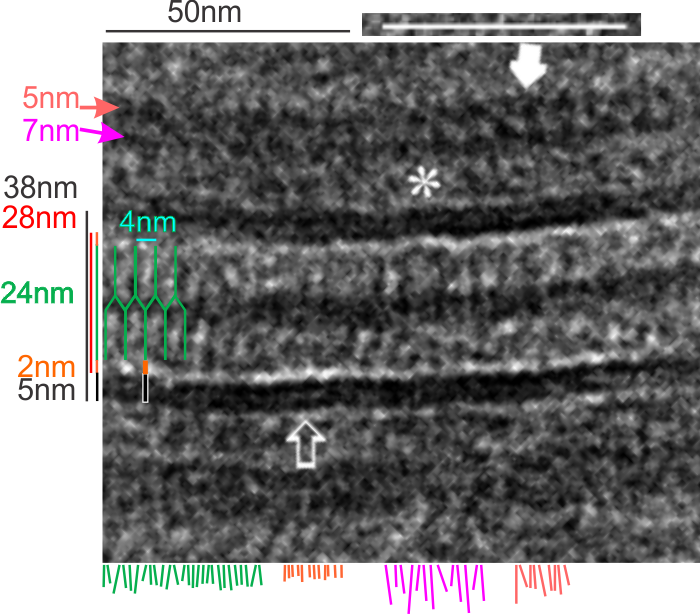
5nm approximate thickness of the cell membrane (one on each side of the desmosome shown here)
38nm from plasmallemma through to plasmalemma of second cell (THEIR MEASUREMENTS LIKELY TO THE CENTER OF THE TRILAMINAR MEMBRANE were about 33nm — pretty close)
28nm extracellular space (THEY DIDN”T MEASURE)
24nm is the @ height of the repeating units and N terminals of the desmoglein and desmocollin molecules (two molecules stacked=height (vertical dimension in this micrograph) (THEY DIDNT MEASURE). It is marked as the green stretchy-wire lines (likely a significant configuration for movement of cells that allows for some “give and take before break” which would not be surprising, but actually be awesome) and was easily drawn over many portions of many micrographs (this, and others of my own that i have posted before) .
4nm spacing between the repeating but alternating units of the desmocollins and desmogleins (THEY THOUGHT THAT THE VERTICAL LINES WERE 5nm APART, I think more like 4nm apart and staggered).
2nm orange lines, the lucent region just before the transmembrane segments of the desmocollins and desmogleins.
7nm pink arrow is a measured periodicity of one band of densities in the inner plaque, and the orange arrow represents measurements from adjacent periodicities. To me they didn’t look the same… the latter perhaps being further spaced and still alternating. White asterisk is from the original micrograph in the Al-Almondi paper, outer white arrow points to the same group of proteins as my orante arrow labeled with 5nm distances. Their white outline arrow points to a lucent area that i did not measure which is just intracellular to the inner leaflet of the plasmalemma.
My measurement lines are shown below the micrograph. Green: (i should have made black since they were measures of plasmalemmal thicknesses).
In liver, the intercellular space between hepatocytes is actually greater than the extracellular space of the desmosome filled with the cadherins…. which really is a very rigorously attached and spaced area.
One other difference in the way that Al Almoudi describes the “inner dense plaque” (most medial area of the intracellular desmosome structure) as having a single periodicity…. but when i counted distances between periodicities they actually were statistically different (p=0.03) with the most medial band of periodicities being further apart than those just adjacent and closer to the plasmalemma (but both in the inner dense plaque.
If i would going to hazard a guess on the shape of the molecules that make up the intercellular space of a desmosome i would have one which has a “blump” before the transmembrane domain, a blump at the N terminal (which is an obvious feature as the central periodic line in the desmosome). The transmembrane part would be quite thin since there appears to be a lucent line just on the outside of the plasmalemma of the two adjacent cells, and before the linear densities are obvious. The current protein databases make an extracellular domain 3-D molecule for desmocollin and desmoglein have a good chance of being fit to the densities found in electron micrographs.


 N
N