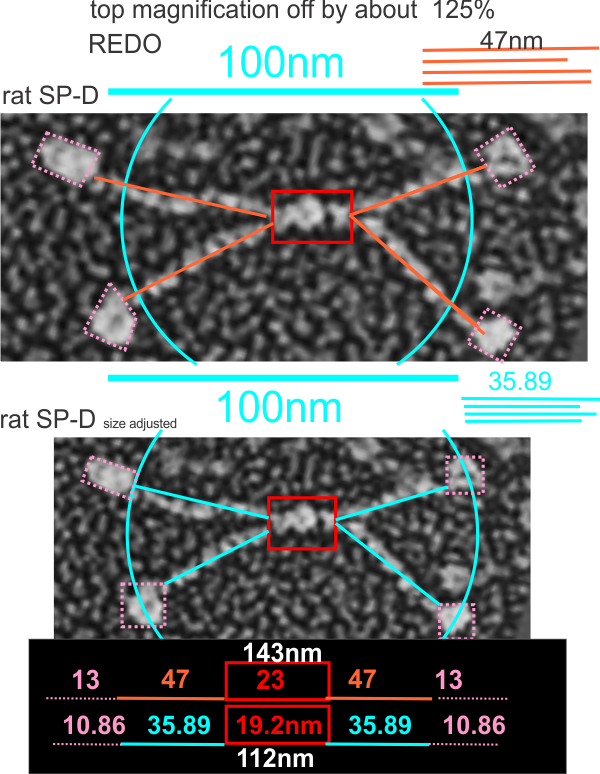
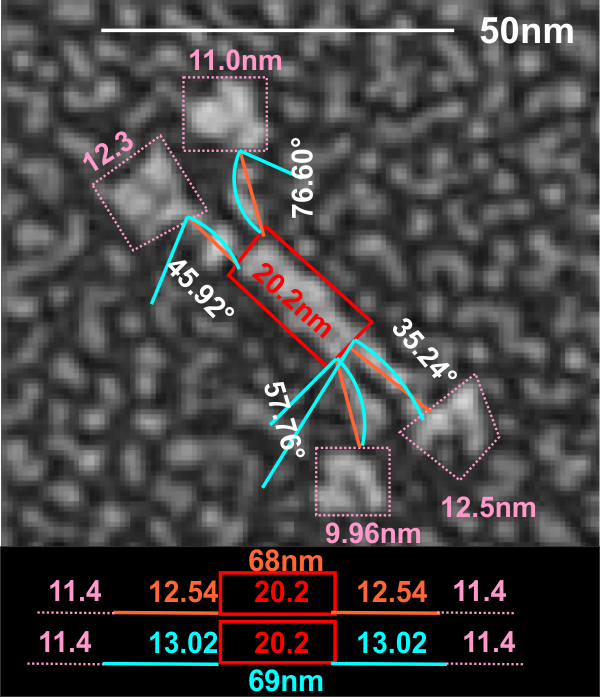
White et al published images of a mini-SP-D which is lacking C3 and C4 exons from the protein. It interests me to examine the differences in curvature and kinks in the arms of the dodecamers and comparisons in length of the arms when those two portions are missing.
It was necessary to figure out what (if anything, i am just looking for answers) was wrong with their micron marker for the full length SP-D and it seems to me that it is off by about 120-125%. This makes comparison with the mini-SP-D difficult. Two different sets of re-calibrations of their image (one with grain size of the adjacent background and one using the oft published size for the full length SP-D dodecamer) are shown below… and this image is a replicate (with different results stemming from the measurement being from the point of crux of the N terminal (probably plus some of the collagen-like domain) to the center of the density that is the neck and CRD. The second set of measures traverses the distance from the crux of the central portion to the edge of the neck and CRD with slightly smaller overall measurements. You can see their original magnification puts SP-D at about 143nm rather than the “just over 100nm” that is consensus in the literature.