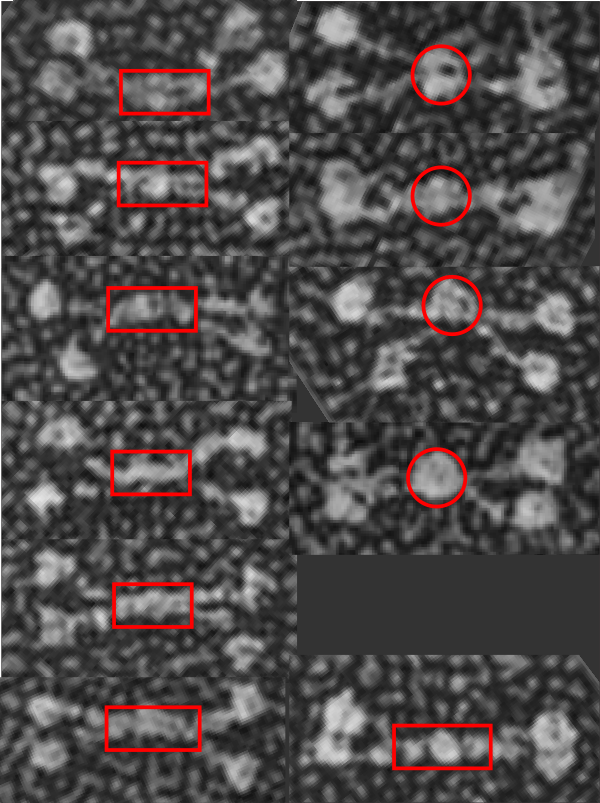
Just a thought here….which came to mind while i swas measuring the arm length and CRD dimensions and N term lenght of 11 mini-SP-D molecules that there was something odd in the center of about half of them. The N terminal of mini SP-D dodecamer (White et al, mentioned frequently) sometimes has an odd look bundled up central portion. I copied and pasted these 11 images to compare them, and 4 out of 11 really did have a greatly bundled (and seemingly) a reduced length. Typical full length SP-D makes a dodecamer with an N terminal (which might also include a portion of the collagen-like domain… as the proportions given in diagrams and in number of amino acids in the N term seem to disparate, and not aligned with the lengths seen in the shadowed electron micrographic images). In particular however, the mini-SP-D with deletions in the collagen like domain show N terminals (about 36% of the time) which are “balled up” into dense areas. Just looking at the layout of 11 molecules below it is also apparent that in those images where the N term areas is balled up, the collagen like sequence (arms) are separate earlier.
In two of the four dodecamers on the right, the CRD of an arm is not clearly defined, there is the possibility that in those cases, the CRD has fallen back on the N terminal. Rejecting that idea, there are also images in the left hand column which have poorly defined CRD and they do not show balled up N terminals. The nicer explanation is that somehow removing the C 3 and C 4 alters the way the N terminals bind together in the center.
That the bar portion in the center of SP-D and mini-SP-D has some of the collagen-like sequence seems
 i think had i been doing this microscopy i might well have been advised to be absolutely precise in my measures of nm length. In retrospect in my own career…. i should also have been more careful.
i think had i been doing this microscopy i might well have been advised to be absolutely precise in my measures of nm length. In retrospect in my own career…. i should also have been more careful.