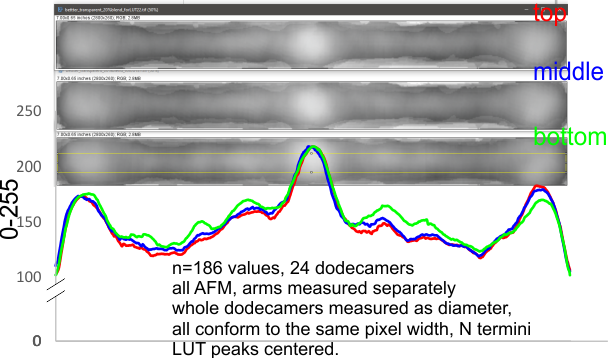
Little change in LUT plots for SP-D dodecamer arms regardless of how I manipulate the image. THis of course gives me some security since it is very unlikely that a change in grayscale, or stretching or shrinking artificially centered N termini peaks and adjusting the arm arcs of the molecules by cutting and aligning and reestablishing arm length according to the nodal (segmented) arm length.
Little change in LUT plots for SP-D dodecamer arms regardless of how I manipulate the image. THis of course gives me some security since it is very unlikely that a change in grayscale, or stretching or shrinking artificially centered N termini peaks and adjusting the arm arcs of the molecules by cutting and aligning and reestablishing arm length according to the nodal (segmented) arm length.
It also is noteworthy that the alignment of 20% transparent images (24 of them – SP-D hexamers, each CRD to CRD section taken as a bilaterally symmetrical molecule (by consensus) does not change the LUT plots either.
It really adds support to the fact that I have not done anything to alter the original image that would jeopardize the basic information.
What is disconcerting is that each set of images (taken from previous publications (Arroyo, Hartschorn, Leth-Larsen, Lu and i will use others for a complete group) show that each of these authors shows micron markers which produce dodecamers that are significantly different in size than the other authors.
Reasonably, shadowed dodecamers are smaller than AFM dodecamers, I can accept that, the techniques are completely different, What I have difficulty accepting is that using images from the same publication (different figures and different micron markers) produce groups of images with significantly different SP-D arm lengths. Each vertical bar on left is significantly different from vertical bar of same color (e.g. pairs).
Girth Sorensen (SP-D Structure, Decomposition, and Proteolytic Degradation –“The amino acid sequence (375 aa) of the mature SP-D monomer consists of four structural domains: (1) an N-terminal domain involved in intermolecular disulfide bond formation; (2) a collagen domain, important for spacing of the CTLDs; (3) an α-helical neck region involved in protein trimerization and spacing of the CTLDs; and (4) a globular C-terminal CTLD (Figure 1A), responsible for Ca2+-dependent binding of microbial ligands. The SP-D protein structure is stabilized by assembly into trimers and multimers via two conserved cysteine residues in the N-terminal domain.”
*= deglycosylated SP-D
** recombinant mutant SP-D with no apparent difference in size (nm arm length) or peak number (LUT table plots)
Some reports of SP-D are dont agree with others.. e.g. The individual “arm” length of an SP-D monomer is approximately 46 nm (therefore under 100nm). But actual arm length may not impact the search for peaks between the N termini peak and the CRD peaks, since I think the best approach is to make them all a uniform length (as done in the images (24 semitransparent)
and plots below using different SP-D transparent images in different stack order to show that the plots are virtually identical no matter how they are organized.