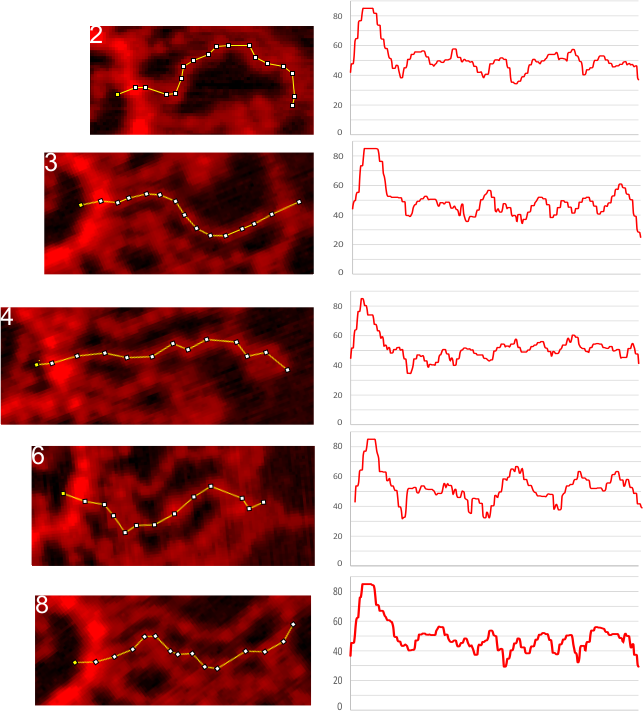
Finding ways to quantify shape, peak heights, widths, and patterns in a molecule which has long arms (100nm+) and lots of flexibility therein has its challenges. PLEASE READ< THIS IS NOT SURFACTANT PROTEIN D and I was NOT prepared by me, but will remain anonymous for the time being. BUT, here is a section of a molecule which has approximately 30 arms with very definite “beads on a string” appearance. Not all of the arms could be traced with sufficient (squinting and mental debating) could be deciphered (in fact relatively few remain untangled enough to produce a LUT plot with any kind of predictability. Here is a stack, which facilitates looking at the arm, the actual ImageJ tracing and the plot of luminance (brighness, — i havn’t figured out which name is the best yet). Cropped and rotated (so that the bright peak is always at the left — and this was the beginning point of the traces, thus the biggest peak is also always at the left) arms are aligned in a column and are all the same magnification and enlargement. The plots at the right are given in the same height as ImageJ scale measured them, but were aligned by nm (100nm) which was a mean length of a trace of several of the straightest and most easily traced arms. No other alignment of the individual peaks or plots was made but picking a “last” peak that is commonly occurring and normalizing that with the beginning of the first peak cold make the interim peaks more easily seen. Hopefully the technique here will prove helpful looking at SP-D fuzzyballs.
There is pretty much a vertical line for a very wide and prominent ‘First peak’ (blue) followed by a regular less prominent peak of lesser height (gray). Many of the arms showed four peaks before a rise to the second tallest middle peak (yellow). Two subsequent peaks show up consistently (green and purple) moving to the right. (TOP IMAGE). Five of the easiest arms to plot also have peaks that line up well (BOTTOM IMAGE panel of 5 arms). (the image used was processed as 2DFFT in gwyddion then opened in ImageJ, and traced with a segmented line)