Still going through all the transmission electron micrographs of alveolar type II cells from 40 years of photography. Here is one print from an owl monkey (no treatment) I pseudocolored with yellow-brown for the nucleus, red for the nuclear pores, blue for the lamellar bodies, cyan (green) for the cytoplasm. Just for the record, i did not find any protein structures within the RER which might be construed as intrcisternal layered RER inclusions that one wants to see if one is looking for SP-A.
Category Archives: Electron microgaphs of lung
Owl monkey type II alveolar pneumocyte
I have searched, with electron microscopy, several different mammalian species for a pattern in the RER of protein which might be considered an accumulation – paracrystalline inclusion like thing comparable to what I have seen in guinea pig, ferret and dog, but none seen so far in owl monkey. Here is a cut and pseudocolored image of a moderately good type II cell from an owl monkey…. taken for another study (anm #1 neg 7160 bl 24060, untreated routine TEM) in 1982. Lamellar bodies are mostly empty in this particular image.
The nucleus is pretty dense, lots of condensed chromatin on the right hand side, and perhaps a portion of the nucleolus is there. There are some dense blue granules (presumed to be glycogen) near the bottom of the type II cell and also scattered elsewhere. (The type II cell was cut out using the eraser tool and lasso tool, and dirt and scratches were removed with the “bandaid” tool in photoshop).
5 year old mongrel dog lung endothelial or alveolar type II epithelial cell? inclusion
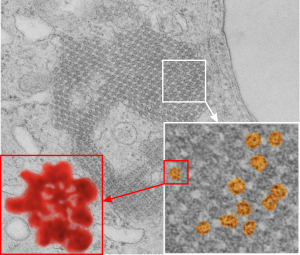
From the previous post, I did some more searching on the very ordered protein inclusion in the type Ii cell of a mongrel dog (5 yr old). The orderly array looked to me like a bunch of hexagonal structures with a central dot. When I made measurements (comparing the magnification, enlargement and pole piece used at the microscope, I figured out what about 500 nm would be, and then 50 nm seemed to be almost exactly twice as big as each of these hexagonal flower (bouquet) structures. They might not actually be octadecamers (as is often found for SP-A )- and a shadowed image of one such shadowed SP-A molecule from a publication by T. Voss, H. Eistetter, KP Schafer in J. Mol. Biol, 1988, 201: 219-227, which i have cut out, pseudocolored in photoshop, cut out, and placed along side one of the bouquets in the ordered protein inclusion in this particular cell. Since I could not identify the cell specifically, one only can guess as to what it might be.
nb. since this blog, I found a similar micrograph of dog lung, in a paper by Stephens, Freeman, Stara and Coffin, Am J Path, 73: 711-726, 1973 which identified a very similar structure (cuboidal organization) in endothelial cells (they say)(Figs 14-16 in their publication). I cannot identify the cell in my micrograph as an endothelial cell, but can confirm that there are dense coated vesicles (at least 11 in this image) just like they found in their micrograph., and one edge of the cell in my micrograph is adjacent to collagen fibers. This would make the cell either epithelial or endothelial. It has no lamellar bodies and no nuclear profile, and only two kind of iff-ish mitochondria.
Circles identify the two molecules (one SP-A according to Voss et al, the other a similar sized protein (could be an octadecamer?? just making a guess here) or some other order of –merization. In the two images (see previous post) there were literally hundreds of times that one could visualize a bouquet with center point as the one enlarged in the inset, and pseudocolored a purple. One of the perplexing things is that when one begins to see structures in a repeated design like this…. many more can be pointed out, and then one wonders what is imagination and what is fact (that is the great tendency of the human brain to become immediately biased).
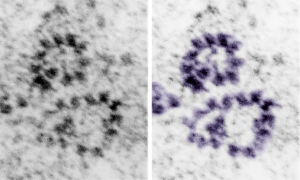
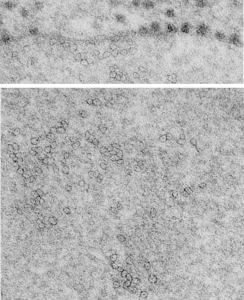
 So after finding this in the publication by Stephens et al, 1973, I decided to scan another area of that same cell and enlarge a portion, and highlight some structures (as above) which look like a multimer of SP-A in a lattice formation and post it here again. Would love to say I could count 18 blobs to depict the carbohydrate recognition domains (haha). I highlighted the “spokes” found in the inclusion below, I should have seen them in the inset inclusion above, but did not highlight them… however, they are there.. maybe 6 of them in top, maybe 7 in bottom inset. The configuration here , globby tops, skinny necks and central dot is pretty reminiscent of how the bouquet of SP-A is diagrammed.
So after finding this in the publication by Stephens et al, 1973, I decided to scan another area of that same cell and enlarge a portion, and highlight some structures (as above) which look like a multimer of SP-A in a lattice formation and post it here again. Would love to say I could count 18 blobs to depict the carbohydrate recognition domains (haha). I highlighted the “spokes” found in the inclusion below, I should have seen them in the inset inclusion above, but did not highlight them… however, they are there.. maybe 6 of them in top, maybe 7 in bottom inset. The configuration here , globby tops, skinny necks and central dot is pretty reminiscent of how the bouquet of SP-A is diagrammed.
RER protein in a type II cell of a guinea pig
The stacks of RER cisternae in this particular type II cell don’t show the layering and patterning that other cisternae from these animals do, that is, they look more like ordinary RER cisternae (though clearly a little dilated, and clearly more stacked). Still I think this is an overproduction of SP-A or some SP as other images from the same animal show the periodicity seen in intracisternal bodies.
The stacks of RER are reminiscent of the stacks of birbeck granules in the Langerhans cells where langrin is overproduced. This is a paper by J. Valladeau et al, Immunity 12: 77-81 (2000) where they transfected langerin cDNA into fibroblasts and got stacks of birbeck granules (which they show in an electron micrograph). While the image below is certainly NOT langerin, and NOT a transfection of any SP-A cDNA into alveolar cells, it is probably the ONLY image that i have found where the profiles or RER might mimic granules (RER) of other c-type lectins (like the birbeck granule) in terms of being segmented and stacked.
Awesome polysomes in a guinea pig type II alveolar cell!
There is no underlying point for this, just was this wonderful coil of polysomes in a type Ii alveolar pneumocyte from a guinea pig and wanted to capture the “beauty”.
Original unedited micrograph (to estimate magnification, each ribosomes is about 20 nm in diameter) is grey scale (L) and a few of the ribosomes and the polysomes are pseudocolored purple (R).
Surfactant protein D rotary shadowed fuzzy ball
Here is an awesome picture in a publication by the following individuals: Martin van Eijk, Chris H.A. van de Lest, Joseph J. Batenburg, Arie B. Vaandrager, Joseph Meschi, Kevan L. Hartshorn, Lambert M.G. van Golde, and Henk P. Haagsman “Porcine Surfactant Protein D Is N-glycosylated in its Carbohydrate Recognition Domain and Is Assembled into Differently Charged Oligomers”, American Journal of Respiratory Cell and Molecular Biology, Vol. 26, No. 6 (2002), pp. 739-747, which I have cropped and repositioned and added a micron marker to.
I am trying to sort out whether the banded protein in the RER of some guinea pig, ferret, and mongrel dog specimens that I have repeatedly observed could be surfactant protein A or perhaps surfactant protein D. It seems to me that It is possible for the 100 nm periodicity with a central dark band to be surfactant protein D, but because of the relative abundance of SP-A in lung, it seems less likely, though not impossible that it is SP-D. Another study by: Stefanie M Heinrich and Matthias Griese Assessment of Surfactant Protein A (SP-A) dependent agglutination BMC Pulmonary MedicineBMC series, shows artificial organization of SP-A, which does the mirror aggregation as I have predicted in my images. They used streptavidin beads in vitro to accomplish this, and while not showing images that I particularly “believed” the diagram they provided was similar to mine.
Upper image the cruciform tetramer, bottom image the SP-D fuzzy ball.
I think SP-D may not be the protein in the intracisternal RER (granules) profiles in guinea pig in my micrographs, because of the low electron density of the space between the CRDs, that similar area in the SP-A showing a greater relative electron density than seen in the fuzzy ball below. This is also more reminiscent of the periodicity seen in the langrin/CD 207 protein in Langerhans cells (Birbeck granules), as you will recall that that linear density is electron pale and very electron dense, not more middle density like the intracisternal protein in guinea pig type II cells in these posts.
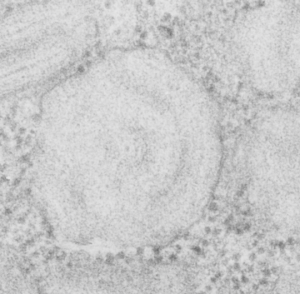
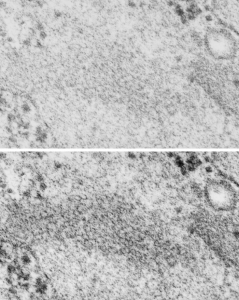
Concentric organization of protein layered in an RER profile presumed to be surfactant protein A
Guinea pig, aged, routine electron microscopy, type II alveolar cell, in particular the RER and a highly organized protein contained: possibly surfactant protein A.
This micrograph is one of the few that is concentric and shows banding at the same time, at least that I have encountered. The center period looks like the others which are only a single set of bands, i.e. a single central dark area, a lighter band, and then the outer band (which would include the CDR part of the protein octadecamers and the RER membrane.
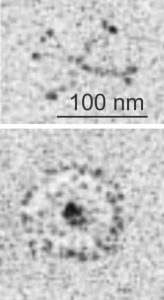
It has ribosomes for size comparison, at 20-30 nm in diameter, and as can be seen from the longitudinal profile of RER in the upper left corner where the banding is more obvious, even in the concentric arrangement, the spacing of a single complete period is about 100 nm. I evened out the exposure of this micrograph, nothing else.
Dark band of RER profile in type II alveolar cell of a guinea pig
Dark band of an intracisternal body (dark bands in a perpendicular section of these intracisternal bodies span about 100 nm, but when spread out over a tangential cut, can have many more nm from the next dark layer in these inclusions. (I am calling them inclusions for want of a better word, but are rather sites of over-production and layered organization of what I think is SP-A. It provides an alternative view of the very orderly periodicity (3,5,7,9 – depending upon orientation and how thin the section is). These two images are identical except that I have increased the contrast, and burned (photoshop) the order which I see in these tangential portions (which likely represent the carbohydrate recognition domains of SP-A. I thought at first these hexagons were SP-A, then began to wonder if i could find similar structures “anywhere” in “any section” as an artifact of fixation, so went to several micrographs and scanned and examined them for randomly occurring small octagonal structures about 25 nm in diameter. There are many, but then I used the “light” banded area just as a control to the dark banding in an RER profile though to contain large amounts of organized SP-A as an internal control, and to help rule out the possibility that the fixation turns all proteins into patterned molecules (which it probably does to some degree) but which would fool one into thinking it was SP-A. So the bottom line is that I think the hexagonal structures (about 20-25 nm in diameter (see ribosomes nearby for comparisons in size at 20 nm).
Guinea pig alveolar type II cell: SP-A bouquets?
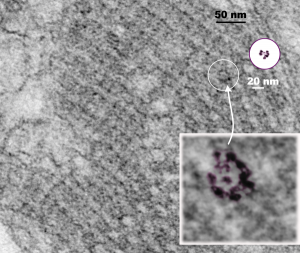
Plugging away at whether these hexagonal structures found within the RER protein accumulations in some alveolar type II cells which are found on tangential sectioning of what I have called “intracisternal bodies” are really SP-A bouquets which are splayed out with a center dot being the neck and collagen portions of that molecule. I have taken a portion of one such intracisternal body which has a small portion which is ribosome studded highlighted (using the burn tool in photoshop) a few of the hexagonal arrangements which seem to have six or so densities on the perimeter, possibly the carbohydrate recognition domain trimers of the SP-A 18-mer.
Previously posted images of those found in ferret type II alveolar cells really looked to be larger, and that is problematic. These hexagonal structures (pictured below) are from an aged, untreated, control, guinea pig and the bouquets seem more in line with the size of a 20 nm ribosome, a string of which I have included for assisting with perception of relative sizes. There is a large tangentially sectioned dark band (actually two dark bands) in the image below unaltered, enlarged with the ribosomes in which I have highlighted the hexagonal molecules (using ONLY THE BURN TOOL in photoshop) to accentuate what I see as patterning. The ultimate test of this would be to take an unrelated micrograph of an unrelated RER inclusion, at the same magnification and see if i could pick out as many hexagons per nm in it as I find in a tangential section of an intracisternal body ( ha ha–sort of pedestrian but it would be a good test).
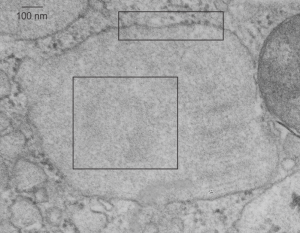
So the upper micrograph gives you a distance marker (bar=100 nm) and the upper box gives you the area of ribosomes and a portion of the intracisternal protein structure which I presume to be SP-A. The lower box gives you the identical area of as is found in the bottom panel, which has two areas of slightly darker electron density marking the tangential portions which are the dark lines of the intracisternal body periodicity. In both enlargements (boxes) I have highlighted just a few of the very many hexagons, which are hopefully the top down view of SP-A molecules.
For those of you who are curious, a mitochondrion is off to the right, and there are also portions of two large intracisternal bodies, and half a dozen or more single – periodicity intracisternal bodies as well. In the central intracisternal body on the right hand side shows the 100 nm periodicity (which on a completely perpendicular section contains anywhere from 3 – 9 bands).
More evidence that SP-A bouquets are found in tangential sections
More evidence that SP-A bouquets are found in tangential sections of the protein filled bodies found in some alveolar type II cells. I am pretty convinced that these bodies are for the greater part surfactant protein A that takes the form of an highly organized (layered) structure that shows linear organization and period repeats that are consistent with an end to end configuration of surfactant protein A, again mirrored. That is, bouquet end to tail end, tail end to bouquet (times 2) per 100 nm period with the central and outside lines of the periodicity representing the carbohydrate recognition domains.
Previous posts show this intracisternal protein on cross section perpendicular to the long axis of the periodicity – or layering, which is easily recognized, and quite orderly, but since the protein inclusions also show curvature and in some cases end to end exchanges in layering direction, and others even as single tubules on cross section, and there are usually “growing ends” with ribosomes along that growing edge of the RER profile. The number of ribosomes counted out comes to something on the order of 2 ribosomes per period, and also there are occasions (shown on this blog site) where ribosomes and the protein (presumably surfactant protein A) appear in the same configuration that one would expect from a top down view of the SP-A bouquet right adjacent to each other, it just seems that the anatomy and the protein structures fit together.
All these new data were found 35 years after the initial discovery of that structure with TEM and that initial search for what this body was, and which started back in 1980 when I queried a respiratory scholar and pediatrician then at CCHMC — who was/is a surfactant specialist — but who also was not interested. So this study was taken off the back burner in 2016 for a little fun and replay. I am quite sure that this RER compilation of protein represents an overproduction of surfactant protein A which behaves in the RER profile like other collectins (langerin being a great example and even showing layering patterns onto which a langrin molecule image can be placed, though producing a different orientation of periodicity than SP-A, but conceptually similar, and obviously related in the greater scheme of the way collectins are ordered within the RER. Here is a quick animation that shows one such tangential section of an RER profile filled with this protein, but the periodicity is spread by a tangential orientation to the plane of the section. The darker regions of the image relate to the darker regions of the banding and those areas represent the top clusters of trimers, assembled as the 18 mer of SP-A open and with a center dot presumably representing the neck. Thee micrograph was only contrast enhanced and the burn tool used in photoshop, two scratches were edited out with the bandaid tool, otherwise pixels were unchanged from the original micrograph scanned at 3600 pixels per inch. Images imported into CorelDRAW, exported as transparent png images, and the animation was made with Swishmax 4. (no data were changed)
Thin sections here are probably about 100 nm thick, maybe a little more, thus there is overlap between one 18 mer and the other. It has to be determined (but looks plausible) that the hexagonal SP-A stacks up like a honey comb.