It is a little difficult to get the whole picture of mitochondrial ultrastructure together in one place and in one image. The the large and complex groups of proteins like ATP synthase, or mDNA and mitoribosomes and the proteins for energy conversion, or calcium storage, ion transport, proteins that are involved in cell growth, division and apoptosis, as well as the basic mitochondrial shape and volume density of and shape of cristae and the amounts of mitochondrial matrix, and many other things of which I am sure i am not aware….are difficult to sort out. Ultrastructural aspects related to these functions are not well understood, and at best poorly diagrammed. I have found one website (biology by the numbers) which does do a great job of labeling size and giving measurements of some aspects of mitochondria, but of course not all, and not neatly organized. There really probably is not going to be much consensus about these variations in mitochondrial ultrastructure because the influence of tissue, cell, metabolic state, types of fixation, the embedment, as well as many variations in scope mag, deliberate manipulation of the images, poorly kept records, presumptions, and the list is endless. So the question is how does one go about diagramming (or imaging) the most educational presentation about the mitochondrion. A really perplexing structure is the “pore” or cristae junctions.
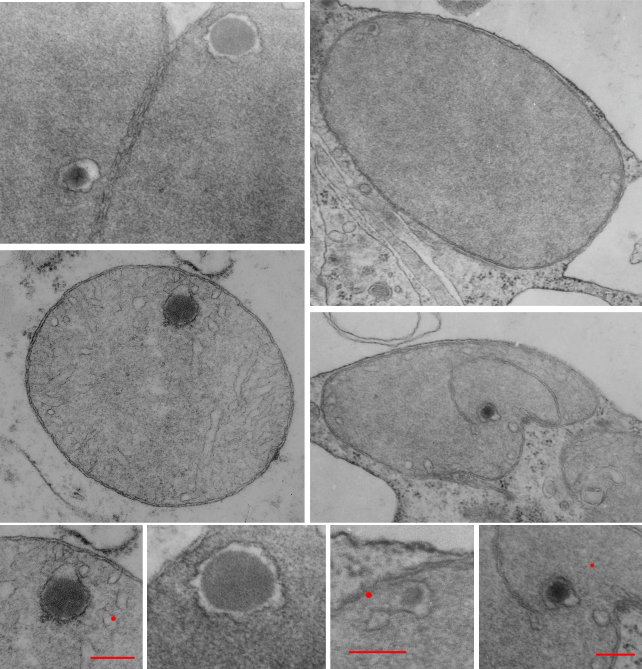
Add to that list of variables, the long list of diseases where mitochondrial shape is found and the concept of imaging the “perfect educational” mitochondrion becomes more difficult. So trying to find a great diagram came about because of an inclusion that I found in the cristae of some of my own micrographs.
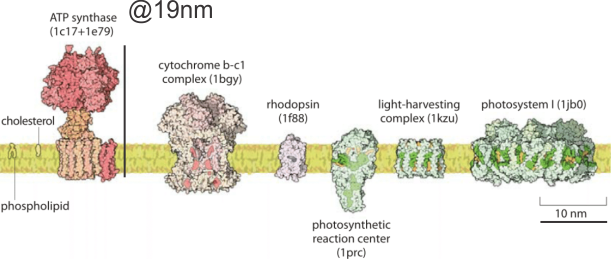
It would just be nice to see these structures ( 5-10nm diameter) in the literature with the normal ultrastructure of the mitochondrion, which is not really that well documented for standard TEM images though there are some nice molecular diagrams of membrane proteins.
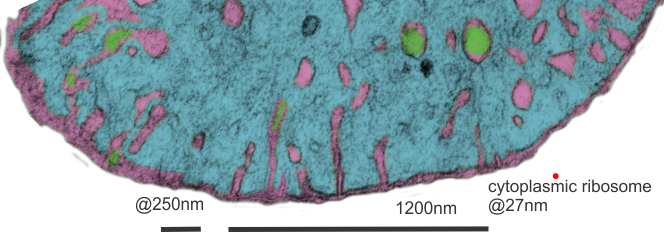
The round electron dense inclusions are unknown (in a crista space)(in these not-perfect micrographs). It is possible that in the bends of a couple of cristae, there are densities right at the bends which would seem to be identifiable. Looking at the blue dots in the top image below, right at the curvature of a crista–one might see those dots as ATP synthase (blue dots are given at the approximate size that ATP synthase should be ( “biologybythenumbers” diagram (thank you for that) (bottom image). Also, separate, larger orange dots could be mitoribosomes which reportedly are just smaller than cytoplasmic ribosomes (picture as red dots (taken from the same micrograph as seen on the left). A second reason for working on mitochondrial ultrastructure is to try to figure out what the electron dense (and homogeneous round) protein is within this crista space)