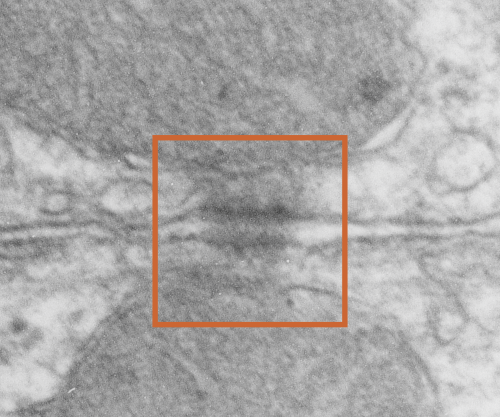
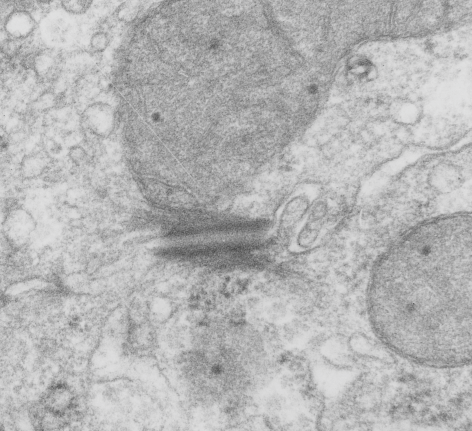
It seems to me that there is a pattern of thickness changes(width or height if you wish… because of the orientation of the diagrams below) and the rigidity of the plasmalemma inherent in the desmosome (likely due to the transmembrane parts of desmoglein and desmocollin ) and that rigidity includes a specific intercellular space dimension. I have seen this published at about 34-38 nm (that is, from the inner leaflet of the plasmalemma of one cell to the inner leaflet of the plasmalemma of the adjacent cell) to be something on the order of 38 nm. Using that dimension, if i measure from inside plasmalemma of one cell to the adjacent cell and compare that to the width from the same places in the 200 nm ring or annulus around the desmosome there is going to be a change in the dimensions (which i could measure as just a few nm greater than that within the area of the desmosome proper. And, then a reduction in the intercellular width (per the more routine proximities of two cells) which becomes something like 40 nm.
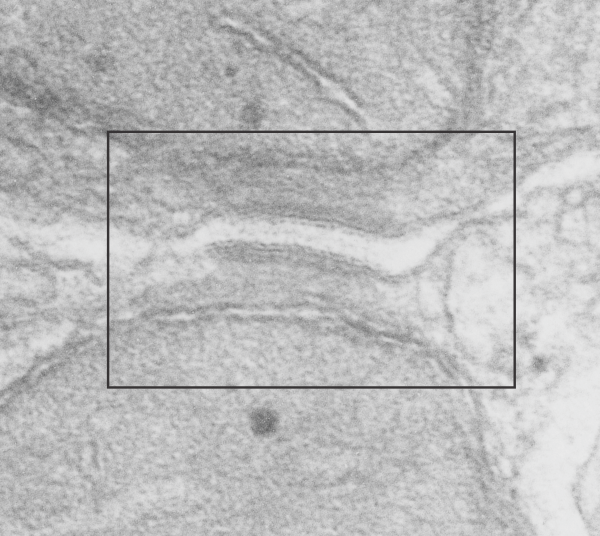
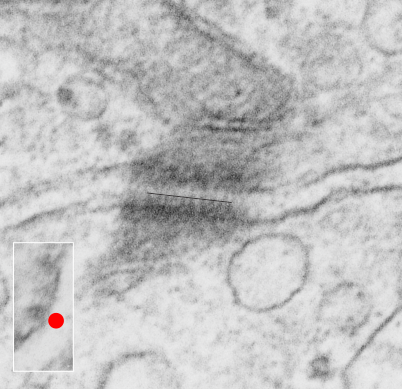
Here is an article which suggests some dimensions for the desmosome, but does not address adjacent variations in the plasmalemmae including the anulus. You can compare with two images and measurements below. Keep in mind that the two images below are not derived completely randomly, as I picked two which had substructure which was clear enough to measure. The annulus of these two desmosomes is indistinct, and longer than that seen in other desmosomes. This likely relates to the plane of section on a perpendicular axis of a round desmosome and surrounding annulus (the latter being increasingly seen as one sections nearer the periphery of the actual desmosome).
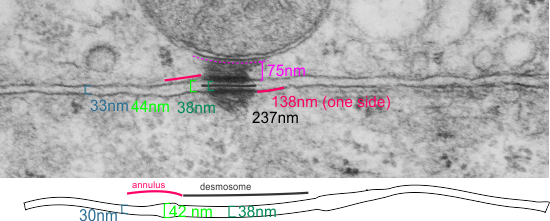
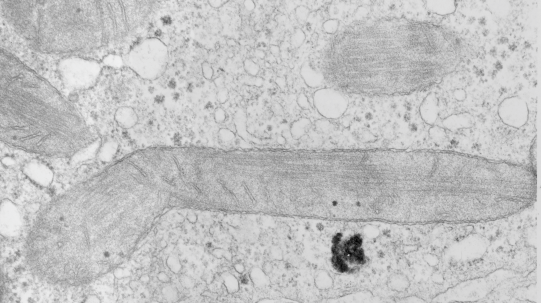
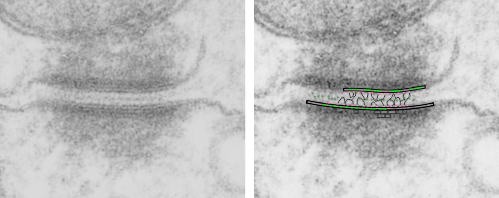
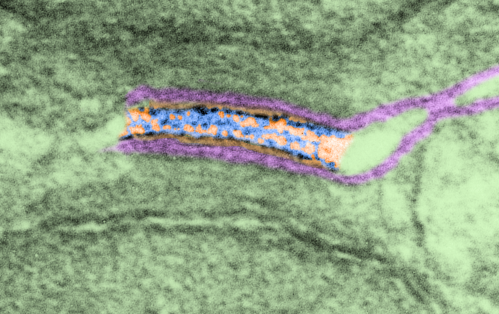
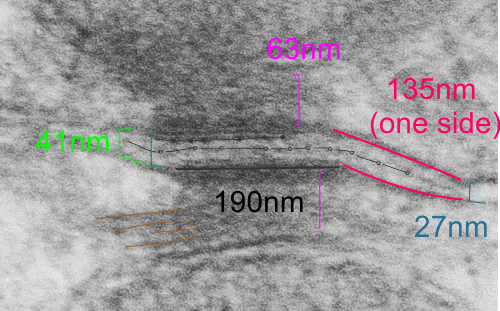 top diagram shows a length of two adjacent plasmalemma, measurements relative to that image, bottom one is a different desmosome, notice brown lines for intermediate filaments, black line for length of the desmosomal spot, dotted lines at periodicities of the bridges between desmocollins and desmogleins, outer dense line with dots, periodicities of the extracellular membrane anchor, pink bracket, thickness of the desmosomal intracellular elements, which includes proteins of the intracellular anchor (desmocollins and desmogleins) plakoglobin, plakophilin, desmoplakin and intermediate filaments (visible at the desmosomal-mitochondrial tether on the bottom more clearly than the desmosomal-mitochondrial tether seen at the top of the lower micrograph. While a big deal is made of the inner and outer dense plaques of the intracellular part of the desmosome, the lower portion of the lower micrograph doesn’t make that case. Were the micrograph sectioned end on to the intermediate filaments below, there might be a more visible inner dense plaque. The outer dense plaque (plakophilins, plakoglobin portion and desmoplakin proteins) is well defined. NB, there is a “flatness” or “rigidity to the outer mitochondrial membrane where the intermediate filaments lie beside it… i hope to search for proteins that might be involved in the linking of mitochondria and intermediate filaments.
top diagram shows a length of two adjacent plasmalemma, measurements relative to that image, bottom one is a different desmosome, notice brown lines for intermediate filaments, black line for length of the desmosomal spot, dotted lines at periodicities of the bridges between desmocollins and desmogleins, outer dense line with dots, periodicities of the extracellular membrane anchor, pink bracket, thickness of the desmosomal intracellular elements, which includes proteins of the intracellular anchor (desmocollins and desmogleins) plakoglobin, plakophilin, desmoplakin and intermediate filaments (visible at the desmosomal-mitochondrial tether on the bottom more clearly than the desmosomal-mitochondrial tether seen at the top of the lower micrograph. While a big deal is made of the inner and outer dense plaques of the intracellular part of the desmosome, the lower portion of the lower micrograph doesn’t make that case. Were the micrograph sectioned end on to the intermediate filaments below, there might be a more visible inner dense plaque. The outer dense plaque (plakophilins, plakoglobin portion and desmoplakin proteins) is well defined. NB, there is a “flatness” or “rigidity to the outer mitochondrial membrane where the intermediate filaments lie beside it… i hope to search for proteins that might be involved in the linking of mitochondria and intermediate filaments.