AFM images (from rhSP-D at pH 7.4 Arroyo et al) of SP-D are informative and numerous and during a careful analysis of them it seems pretty obvious that there are a significant number of occasions where there is a close “sticking” together of the strands of the trimer from the N termini junction, through the tiny peak on either side of the N peak and up to the glycosylation (often including it) peak(s)(plural here because there are a significant number of imaging and signal processing applications that count more than one peak in the area considered the glycosylation peak).
Here are two images (labels on each show the number of nm diameter, the length in nm of each of the hexamers, and arrows that show where the trimers from two hexamers are in close association. I scanned 83 images from the images of Arroyo et al, and found that such an association (which in order to be visualized with any confidence requires that the dodecamer to be lying such that the rest of the trimer-arms are separated, thus not just overlapping — overlapping trimer arms were not included in the count of closely associated N, tiny peak, and glycosylation peak associations.
I have also given a number (my reference number for the set of thumbnail images, and the set of dodecamers to which some measurements have been applied.
Any reason for such a close association between trimers in that specific location is not known by me, comments welcome. 100 nm bar is given for your “enjoyment”, green nm measurements correspond to green segmented tracing through the center of the hexamer to which it refers, same for red segmented line and nm values. Diameter values (orange) are made in ImageJ, and requires that three of the four carbohydrate recognition domains be touched. It is easy to see that once past the glycosylation peaks, the arms of each trimer are separate. On the bottom image, the trimers are close on the top half of the micrograph, but not the bottom half, as the glycosylation peaks are separate. (42 dodecamers, of 86 total, show N, tiny peak, glycosylation peak closeness on one or both sides of the dodecamer.)
Category Archives: surfactant proteins A and D
Four dodecamers: peak(s) 8 width – carbohydrate recognition domain
THis peak has the closest to registering two peak(s) within the larger element of a whole CRD peak area.

THis peak width (just under 17 nm is close to what has been seen before.
Number of peaks within the CRD whole peak depends clearly upon the path of the segmented line used to trace that end of the SP-D trimer.

Four SP-D molecules: Sixth peak(s) and sub-peaks
Same process for determining peak width, and sub peak number…. as before. peak 6 i expected to show up a little wider (here it is about 7.5nm+0.12). in previous plots that are colored one can see it as that peak(s) which are white.

 summary plot from months ago, below to remind one what the scheme of the bilateral symmetry was/is.
summary plot from months ago, below to remind one what the scheme of the bilateral symmetry was/is.

PEAKS PER TRIMER (trimer=about 74 nm including the full N term peak with each plot)
Peak 1=N term peak (full) (peach) @20nm
Peak 2=Tiny peak (purple) @2.5nm
Peak 3=glycosylation peak(s) light green @8nm
Peak 4= unnamed (darker green) @12nm
Peak 5= unnamed (pink) @4nm
Peak 6- unnamed (white) @7.5nm
Peak 7= Neck (yellow) @4.5nm
Peak 8=carbohydrate recognition domain peaks (orange) @ 17nm
Four SP-D molecules: Fifth peaks – lateral to the fourth peak(s) which are lateral to the glycosylation peaks in this bilaterally symmetrical hexamer
This is a small peak by comparison to the glycosylation peak(s) and to the fourth peak just lateral (in a bilaterally symmetrical hexamer) which is @12nm in width, this very narrow and lower peak is only 4nm wide (at the valleys). In previous ridge (Joy) plots it is colored in a bright pink just in case you want to look up its position and relative height and width.
After finding width and number of sub-peaks for each of the 7 peaks on either side of the N, i will make a diagram with the dimensions (width, height, and sub-peaks) found for each of the four dodecamers in this short list of test molecules.
There is a peak 5 in 74% of the trimers, less than 1 per trimer, and no sub-peaks noted.
Four SP-D molecules: Fourth peaks – lateral to the glycosylation peaks
Here is a peak series that I think occurrs with a frequency that legitimizes it as being thought of as a distinct entity. The peak number in grayscale plots for SP-D hexamers has been calculated by numerous signal and image programs but the pattern itself is the result of observing countless images and the patterns within each hexamer mirrored as two trimers. (N being central (peak 1), tiny peak lateral to N (peak 2), glycosylation peak being (3), and this peak (4) with unknown function or quality is consistently as wide, with a lower peak height than the glycosylation peak. The divisions are mine. It appears (looking at four dodecamers of SP-D (as individual trimers) to be about 11 nm in width. It does show subpeaks just like the N and the glycosylation peak. It would be amazing if the subpeaks were indicators of the “twisting” together at a subtle distance the three individual SP-D molecules. Thats not too far fetched. This conjecture might hold up until the CRD are reached which becomes rather random positioning of the globular part of the protein.
It helps to visualize this peak (deep green in the ridge plots link to one here). The “tiny peak” (purple) can be viewer towards the top of this ridge plot, but not at the base of the plot.
The number of sub-peaks (tabulated without bias within the five signal processing peak finding algorithms, and the hand counts are consistent with this peak being one for sure but maybe with a smaller element or subpeak. I am defining sub-peaks as smaller peaks which visually appear to be part of a single wider taller peak.
Four SP-D molecules: 13, 14, 15, 16, 17? peaks per hexamer
n=4 dodecamers, with many different signal and image processing filters, giving rise to about 114 different plots and peak numbers; the peak number summary of each dodecamer provides: n=4, 17.25 total peaks per hexamer+2.48. This is still just four molecules.
1) N term peak = @20nm which is divided into two sub peaks about 50% of the time.
2) Tiny peak of @3nm either side of the N term peak
3) Glycosylation peak @9nm, either side of N term peak, and consistently composed of two parts (two peaks)
Four SP-D molecules: glycosylation site peaks within peak
Using just four SP-D molecules, with arms plotted as hexamers, and recorded as trimers (reversing the mirror symmetry and using the N term value as the whole N term peak width), the glycosylation peak (now called the 3rd set of peak(s) in the trimer) sometimes has a lumpy appearance, and signal processing programs routinely divide that single (sort of) largest of peaks (second to the N term and the CRD peaks) into separate adjacent but clearly contiguous peaks, sometimes as many as 6. When i see the brightness areas i dont often agree on what the signal processing programs call “separate” peaks as they really come together in one single significant elevation, clumped as separate but connected peaks. The number of separate peaks within what is typically pointed out as the glycosylation peak in AFM micrographs actually has more than one peak… these data sort out using 4 dodecamers (16 trimer plots – assessed by 5 signal processing programs, several imaging processing filters, and one unprocessed image collectively) that about 2 peaks show up in the glycosylation “bright” spot. 1.92+0.204. This will likely change with the addition of more molecules added to the set.
Four SP-D molecules: glycosylation site peak width
Below are the numbers for peak width for the glycosylation peak. The characteristics of peak, that is whether it is a single simple peak or a peak with multiple elements, I dont find good mention of in the literature.
It is worth noting that because there presumably are three sites (one per strand of the trimer) which are potentially glycosylated, and whether there could be a bumpiness to the bright areas in an image representing the glycosylation peak. This might reflect structural features of “image” the three single strands if they are or are not glycosylated while wound in to a trimer thus causing a displacement of glycosylation sites perhaps showing up as peaks within the glycosylation peak (envision three beads on three separate cords, wound tightly, each displaced slightly creating a lumpy look. This is completely different than the images which show the large CRD and neck elements which do show large bright areas quite distinctly separated at either end of the hexamer. see CRD peak appearance with AFM images, left, and possible completely glycosylated trimer on right (NOT TO SIZE).
In most micrographs there is a dominant “bright” region which has been identified as the glycosylation peak in numerous publications, including the one by Arryoy et al which i rely upon heavily) as a single peak. Close visual examination reveals a couple of things, 1) it is a peak of varying width, and 2) sometimes it is possible to see small sub peaks within the whole. Signal processing and peak counts from images before and/or with the application of image filters, it is common to see bright spots within what might be considered a single larger bright region (brightness in this context is higher on the gray scale value y axis than lower). Defining the “modest but colvincing ” number for peaks along a trimer has been the focus of this research. Both total relative peak heights and valley to valley widths (not taking into account slope or mid peak areas) have yet to be determined for a hexamer of SP-D.
So using just a half dozen analysis protocols on FOUR (4) SP-D dodecamers (16 trimers – where the entire N term peak is included in each trimer), the first three peaks (beginning at the whole N termini junction peak) looks like this. N=19.86nm+1.46, tiny peak=2.53+0.74, glycosylation peak width=8.4+0.69.
My tendency was/is to see the AFM images of this molecule as having 15 peaks. Two pominent CRD peaks at either end of the hexamer, one peak (possibly with its own central valley (so moving the odd number of peaks to an even 16) in the middle and two recognized peaks either side of the N term. These peaks are numbered in many of these blog posts as Peak 1=N term, two tiny peaks on either side of 1, mirrored as peaks=2, then the two glycosylation peaks =3 (either side and lateral to the tiny peaks and sometimes clustered peaks. Peaks which follow I am now suggesting are “real events along the collagen like domain” as peak 4, moderate size, fairly wide peaks, next very narrow low peaks=5, broad low peaks=6, and sometimes visible peak which appears to be the coiled coiled neck domain where sometimes it is covered by the CRD, sometimes not=7, at the end, the CRD peaks which are very prominent sometimes exhitibing clustered peaks=8. These I have colored with specific colors through out the peak counting process, as my own assessment of which of these categories the signal processing peak programs miss the mark (which sometimes they do, sometimes they dont).
Not all trimers show all peaks, thus the attempt to find a mean width, peak height and humber amont the dozens of AFM images I have collected.
Here is a link to description of the ridge plot (Joy plot) shown below.
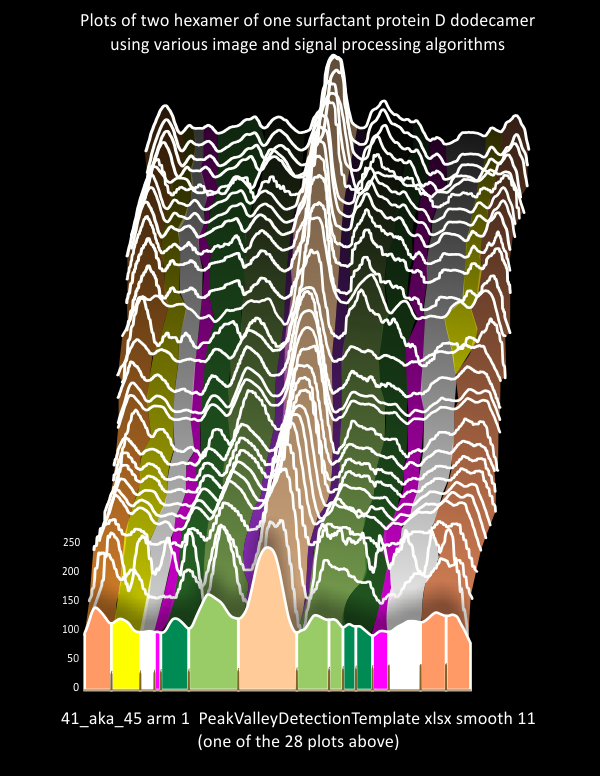
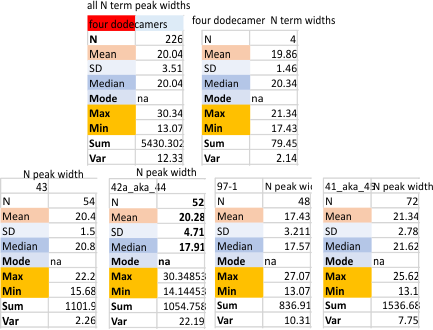
Four surfactant protein D dodecamers: N termini peak widths
Summary of peak widths (valley to valley) using image processing as well as five signal processing programs to calculate that dimension. In one sense the results are really good, in another sense they are basically so similar to those just done by “my minds eye and a good measuring stick” that the two years has provided no new information. 20 nm is a very stable number for the valley to valley measurement of the N termini junction of surfactant protein D as is shown in AFM image. This value is found as all trimers with the full N term peak, as for each of the individual molecules that have been measured (in this case my numbers for them are arbitrary: 41_aka_45; 42a_aka_44; 43; and 97-1) and also the mean of the four dodecamers (N=4) . The number of trimers measured in one molecule was greater than the other three so this data set was primarily made only those plots which have similar processing. That is with 1: images with no processing, 2: images with a gaussian blur (either 5 or 10px); images that have a gaussian blur and a limitrange filter. 3: each of those images from each of those groups of image processing were then exposed to signal processing by five different algorithms (as part of several programs: Octave (Autofindpeaks.m, and iPeak.m; Scipy; LagThresholdInfluence (batch processing) and an excel template for Peakand ValleyDetectionTemplate.xlsx). Therefore the balance of bias was similar among the four images of SP-D.
 The tiny peaks on either side near the valley of the N term peak are located by image and by a combination of image and signal processing algorithms only about 31% of the time. This is a little disappointing, but hand counting the peaks provides a more robust counting of those tiny but very consistently found peaks.
The tiny peaks on either side near the valley of the N term peak are located by image and by a combination of image and signal processing algorithms only about 31% of the time. This is a little disappointing, but hand counting the peaks provides a more robust counting of those tiny but very consistently found peaks.
The number of peaks WITHIN the N term peak varies from 1-2 typically. About 50% of the N term peaks consist of two identifiable sub-peaks. The widths of the sub-peaks is not always equal and depends upon how the segmented line is drawn through the image.
Width of the peaks is about 3nm wide, which still can be identified within the micrographs very often. In this set there is one value which is large, skewing the data. 
I calculated that single image without the large value below.
 Here is the information (peak width in nm of the tiny peak) from months and months ago, which literally has not changed with the addition of the signal processing assessments. I dont know whether to be happy this didn’t change, or be miffed because of the amount of time to confirm that the original assessment was pretty good. Tiny peaks appear about 30% of the time (rather are measured as “peaks” by the five signal processing and one image processing plots about 30% of the time. There are many times that tiny peaks are visible, just are not counted.
Here is the information (peak width in nm of the tiny peak) from months and months ago, which literally has not changed with the addition of the signal processing assessments. I dont know whether to be happy this didn’t change, or be miffed because of the amount of time to confirm that the original assessment was pretty good. Tiny peaks appear about 30% of the time (rather are measured as “peaks” by the five signal processing and one image processing plots about 30% of the time. There are many times that tiny peaks are visible, just are not counted.
)
Four surfactant protein D dodecamers: trimer length in nm
The length of a trimer was determined from the center of the N termini junction peak to the edge of the carbohydrate domain (since this is a lumpy molecule…. the most central route for the line (plot) through the CRD was used. Many examples of the type of line are given in previous posts.
Applications were AS BEFORE- No processing, gaussian blur (5 or 10px) and gaussian blur with limitrange (100 (or above)-255), and each is then subjected to all of the following signal processing applications for peak detection: Lag 5, Threshold 1, Influence 0.05 (batch process), Scipy (Prominence 2, Distance 30, Width 5, Threshold -, Height -), Octave, autofindpeaksplotx,y and iPeaksM80, and excel template PVDTxlsx smooth 11. This means that there are two hexamers (four trimers) each with no processing, some processing, and each of the latter subjected to 5 different signal processing routines. Some select other processing can be included.
Trimer widths in nm, as all lumped values, individual values, and as a single group of four.
The number of peaks per trimer is calculated with all independent measures, as molecule measures (that would be each trimer of four individual SP-D images). The N term peak is calculated in full, WITH each N term peak that is not divided into two peaks. So adding up the peak number — keep in mind that N is counted twice if it is a single large peak.
 Previously, using dozens of image and signal processing programs for literally hundreds of plots, of a single SP-D image (41_aka_45), the number of peaks per trimer was 8. Check out the post here. It is very encouraging to have a selected set of image and signal processing programs provide almost identical results to that original single molecule dataset. That means to me that the gaussian blur and limit range imaging filters can be used somewhat confidently to provide easier counting of peaks along an arm of a hexamer (or trimer) of SP-D.
Previously, using dozens of image and signal processing programs for literally hundreds of plots, of a single SP-D image (41_aka_45), the number of peaks per trimer was 8. Check out the post here. It is very encouraging to have a selected set of image and signal processing programs provide almost identical results to that original single molecule dataset. That means to me that the gaussian blur and limit range imaging filters can be used somewhat confidently to provide easier counting of peaks along an arm of a hexamer (or trimer) of SP-D.
Just analyzing the left hand side of each of the four images, the trimer length in nm is different (as relates most likely to preparation artifact, in stretching or folding of the arms.
 just the righ hand side of the image (the second trimer in the hexamers to be traced are as follows.
just the righ hand side of the image (the second trimer in the hexamers to be traced are as follows.
 There are so many ways to sum these arms up i just decided to do them separately as 16 trimers comprising 4 dodecamers. It makes little difference that i can detect and a nice conservative number of 145 nm as the usual hexamer length, and 73 for the usual trimer length (not exact yes… ). but counting more molecules might be more efficacious than deliberating on just four.
There are so many ways to sum these arms up i just decided to do them separately as 16 trimers comprising 4 dodecamers. It makes little difference that i can detect and a nice conservative number of 145 nm as the usual hexamer length, and 73 for the usual trimer length (not exact yes… ). but counting more molecules might be more efficacious than deliberating on just four.
Two conclusions. 1) Image processing is helpful, signal processing to count peaks, not so much. The plots smoothed with the gaussian blur and enhanced with the limit range function are easy enough to manually count peaks, and the peaks counted manually are guided by ones knowledge which the current algorighms are not (as in not recognizing symmetry, and not permitting small peaks to follow big ones. etc). When a few more molecules are counted and added to the list then perhaps I will find someone who knows how to “train” peak counting algorighms. LOL.
2) The number of peaks per hexamer is likely to be between 13 and 15. THis is much greater than that proposed by Arroyo et al, who found 5.