When examining protein structure with TEM it doesn’t escape me how the patterns sometimes fit into these very regular but seemingly not relevant patterns. In looking at the densities which appear in the center portion of the desmosome and also the diameter of cross sectional cuts of the intermediate filaments associated with desmosomes, i get confused as to why both look to be about 10nm.
I have tried to google such phrases as “texture of lead citrate staining” with TEM, or uranyl acetate or osmium textoxide, or some other texture analysis that might put a “cap” on the limits of where one can go when measuring protein patterns with TEM.
Any images that I review for protein patterns I am forced to find adjacent areas and see if similar patterns occur.
It makes sense that fixation conditions will greatly influence the final texture of the tissue, and I doubt whether there have been any well controlled morphometric studies comparing the myriad methods for processing tissues for TEM. I found an article bu R. W. Horn on this and here is the first two lines as as quote “For the practical biologist applying electron microscopy to the study of biological macromolecules, there are serious problems in obtaining high resolution images showing detail below 2.5–3.0nm. The limitation in resolution from biological specimens can be attributed to support film thickness and granularity, specimen preparation, irradiation damage, focusing effects and possible contamination in the electron beam.” which i will not read. (Somehow it seems a travesty that Wiley (Bruker) think that I should pay to read a pdf the data for which were mostly like funded with public funds — i wont pay). But some terminology and also the recognition of 3nm being an important “stop” point beyond which any assessment of rhythm or pattern in a micrograph would just be “gestimate” is reasonable. I actually, at least for my micrographs, probably from negatives not developed with the most fine grain developer (a penny wise and pound foolish choice made in my career to not use the most fine grain developers for my negatives and prints…I was at that time unable to think to the future of microscopy and structural protein biology and the links possible between the two fields, so sad really).
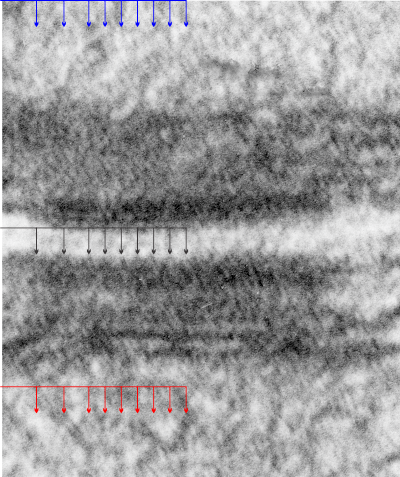
So here is a micrograph of a mouse hepatocyte desmosome which is cut tangentially, the central dense line where the desmocollin and desmoglein come together (but it is still slightly visible), but there areas where the latter two molecules traverse the plasmalemma, there is a little density… at least I think it is periodic. I have added black arrows to indicate where i see those periodicities in the outer lamina of the trilaminar membrane, and moved that same set of arrows to different sites in the micrograph (in this case both are intra-mitochodrial) to see if by chance the same periodicity might arise (thus helping to convince myself that the periodicity actually means something). You can see that the red and blue arrows dont line up exactly with any matrix proteins in the mitochondria.