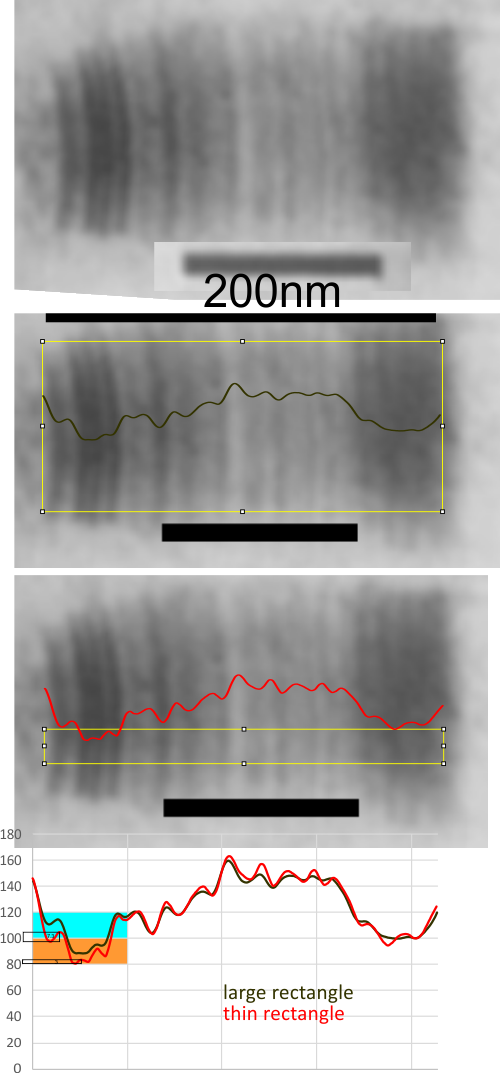
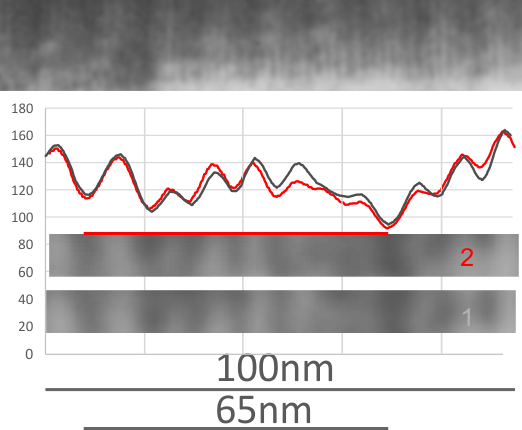
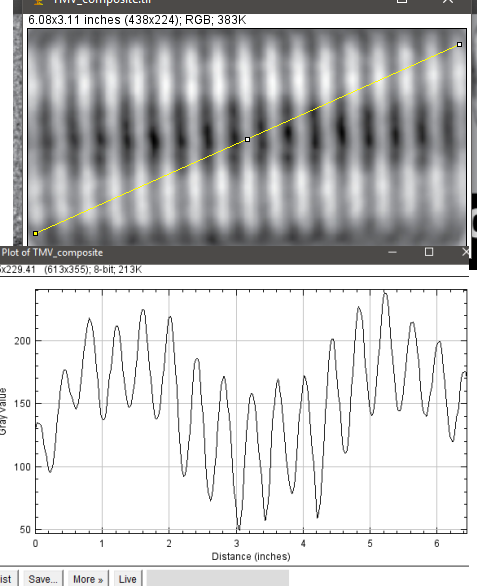
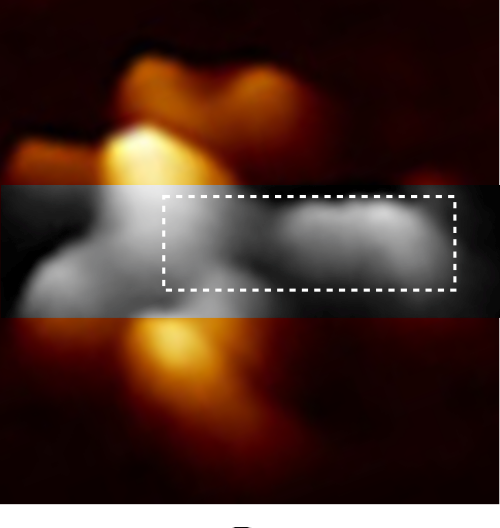
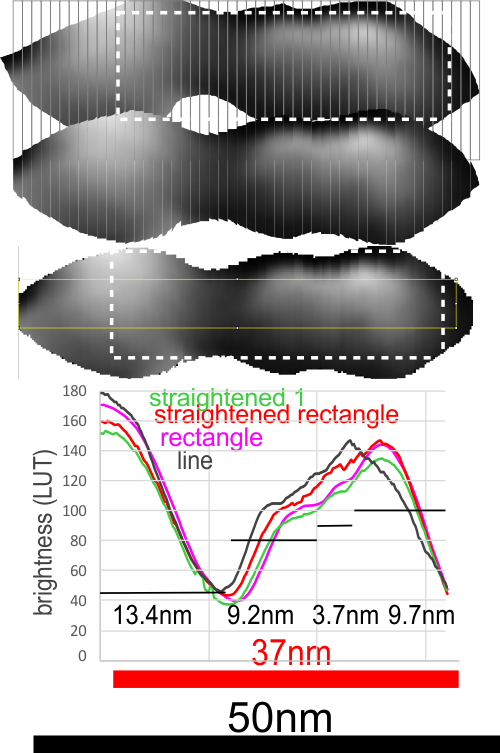
Geometric values all related to DNA according to wikipedia, and a cute hand drawn diagram.
For each base pair, considered relative to its predecessor, there are the following base pair geometries to consider:
Shear: a force acting in a direction parallel to a surface
Stretch: a one-way stretch
Stagger: an eclipsed conformation is a conformation in which X and Y on adjacent atoms A, B are in closest proximity, implying that torsion angle X–A–B–Y is 0°.
Buckle:
Propeller: rotation of one base with respect to the other in the same base pair.
Opening:
Shift: displacement along an axis in the base-pair plane perpendicular to the first, directed from the minor to the major groove.
Slide: displacement along an axis in the plane of the base pair directed from one strand to the other.
Rise: displacement along the helix axis.
Tilt: rotation around the shift axis.
Roll: rotation around the slide axis.
Twist: rotation around the rise axis.
x-displacement:
y-displacement:
Inclination:
Tip:
Pitch: the height per complete turn of the helix.
I will work on a diagram that is an improvement over the one found on wikipedia.