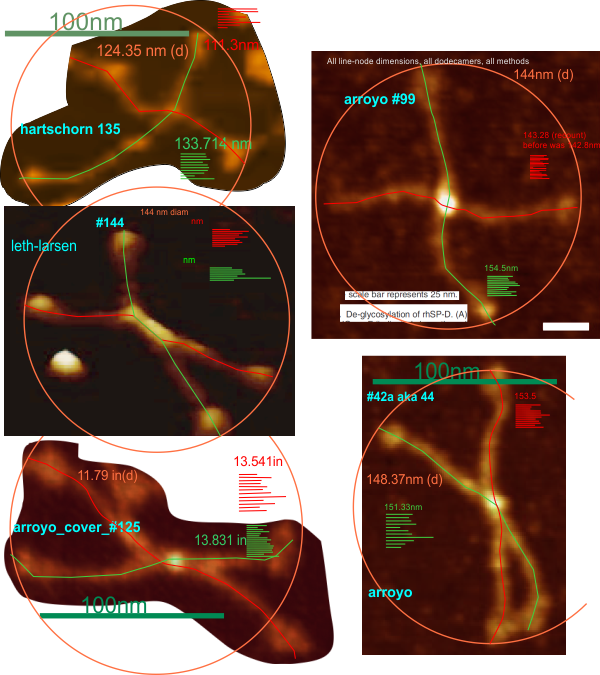
60 SP-D dodecamers from three publications, two authors and a cover image have been measured to find an “arm length” for the separate arms of each molecule, that is, one measurement for two trimers N termini attached, two of these per molecule, again that is from one CRD to the opposite CRD is one set of trimers, and there are two of these per dodecamer which have been measured with a line which has nodes (cusps) and therefore represents a “straightened” length, not just a “crow fly” length. 120 measures from those 60 molecules.
Another set of measures, also totaling 60, was obtained by using a diameter from a circle which touches at least 3 of the four CRD. These two types of measures are not statistically different… however it is close, the linear measures of arm lengths being just a little greater than the diameter.
Arroyo et al images have been used from their Fig 1, Fig 3, Fig 6a, and from their cover (wherein lies the greatest source of variability in size since I believe the mentioned micron dimensions are in error. Additionally, there are small variations in the general size of SP-D dodecamers from Figure to Figure. ONE molecule is de-glycosylated so there are two arms measured with the line which cusps to accommodate the curvature of the arms, which was very close to the mean anyway, thus included in the analysis.
Images (albeit not as nice as those from the Arroyo et al paper) from the paper by Hartschorn et al were measured as well and that mean was smaller (18nm) than the overall mean (of the 120 arms and the diameter measurements) while the cover images of Arroyo et al were about 25nm larger than the mean.
N1: 120
df1 = N – 1 = 120 – 1 = 119
M1: 148.4
SS1: 63382.63
s21 = SS1/(N – 1) = 63382.63/(120-1) = 532.63
Treatment 2
N2: 60
df2 = N – 1 = 60 – 1 = 59
M2: 143.05
SS2: 26472.99
s22 = SS2/(N – 1) = 26472.99/(60-1) = 448.69
T-value Calculation
s2p = ((df1/(df1 + df2)) * s21) + ((df2/(df2 + df2)) * s22) = ((119/178) * 532.63) + ((59/178) * 448.69) = 504.81
s2M1 = s2p/N1 = 504.81/120 = 4.21
s2M2 = s2p/N2 = 504.81/60 = 8.41
t = (M1 – M2)/√(s2M1 + s2M2) = 5.35/√12.62 = 1.5
The two tailed t-value is 1.5047. The p-value is .134173. The result is not significant at p < .05