What I have experienced in 55 years of microscopy is that I like what my eyes see (with my mind’s eye) better than I like what i can “quantify” using the tools currently at my disposal to tell me whether what I see is significant or not.
In times past, the tissues were logged with numbers (after 100,000 blocks I certainly could not remember a number and animal treatment group) the sections, slides, the grids, the stains and the order presented were kept “blind”. The data were talled according to the random choice of image in a random choice of slides or grids examined, all in an heroic attempt NOT to be biased. And why? Bias is easy to insert. Unknowingly inserting bias leads to significant error in science. Bias is inserted both “for and against” the predicted results, and either direction damages the credibility of the data.
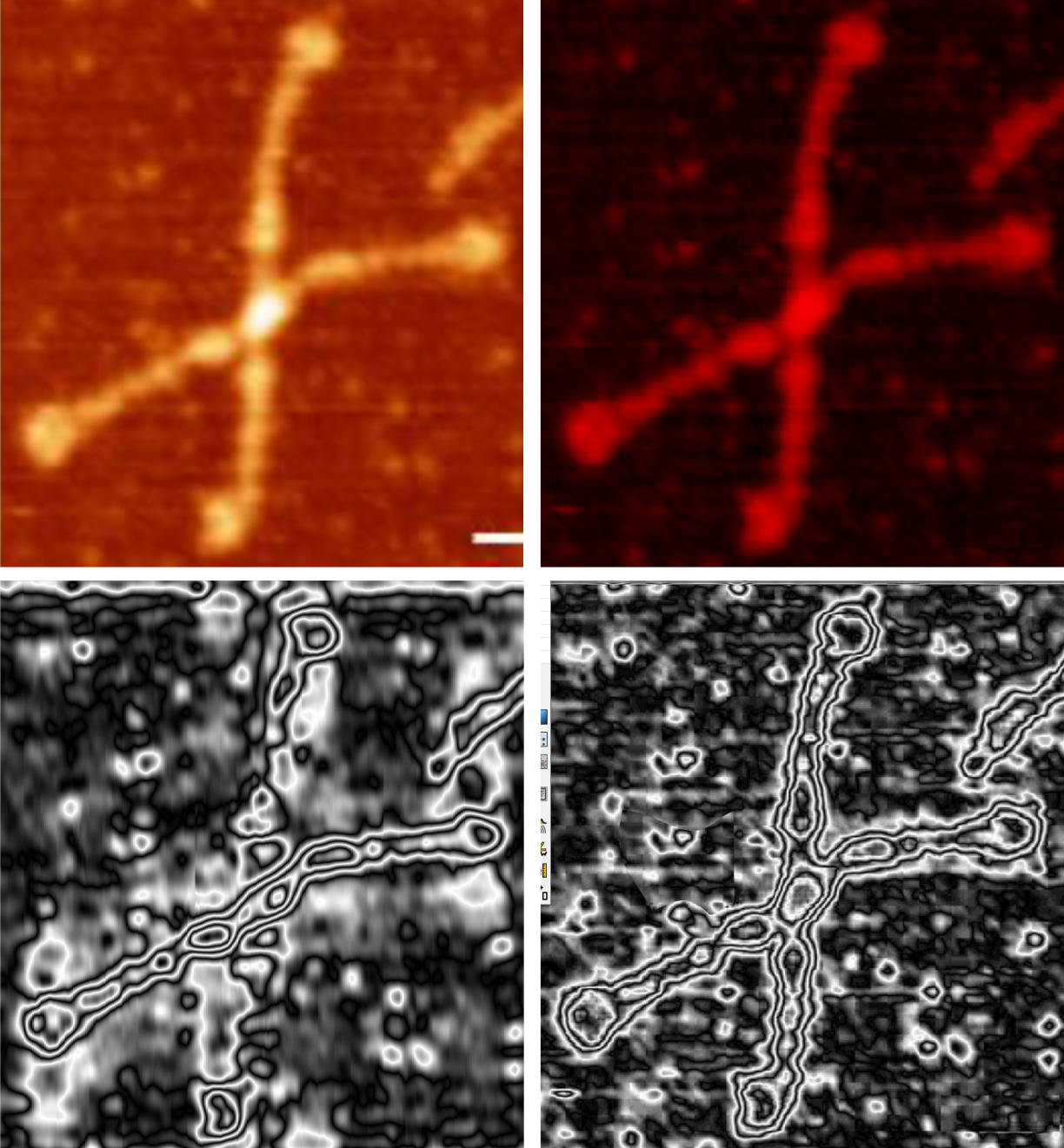
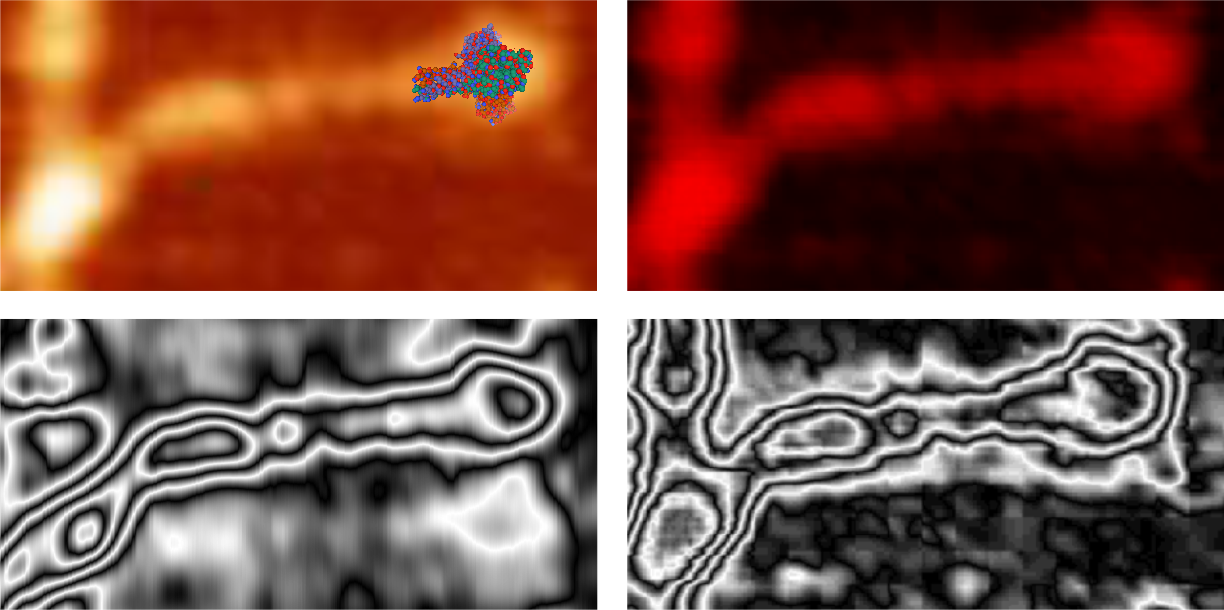
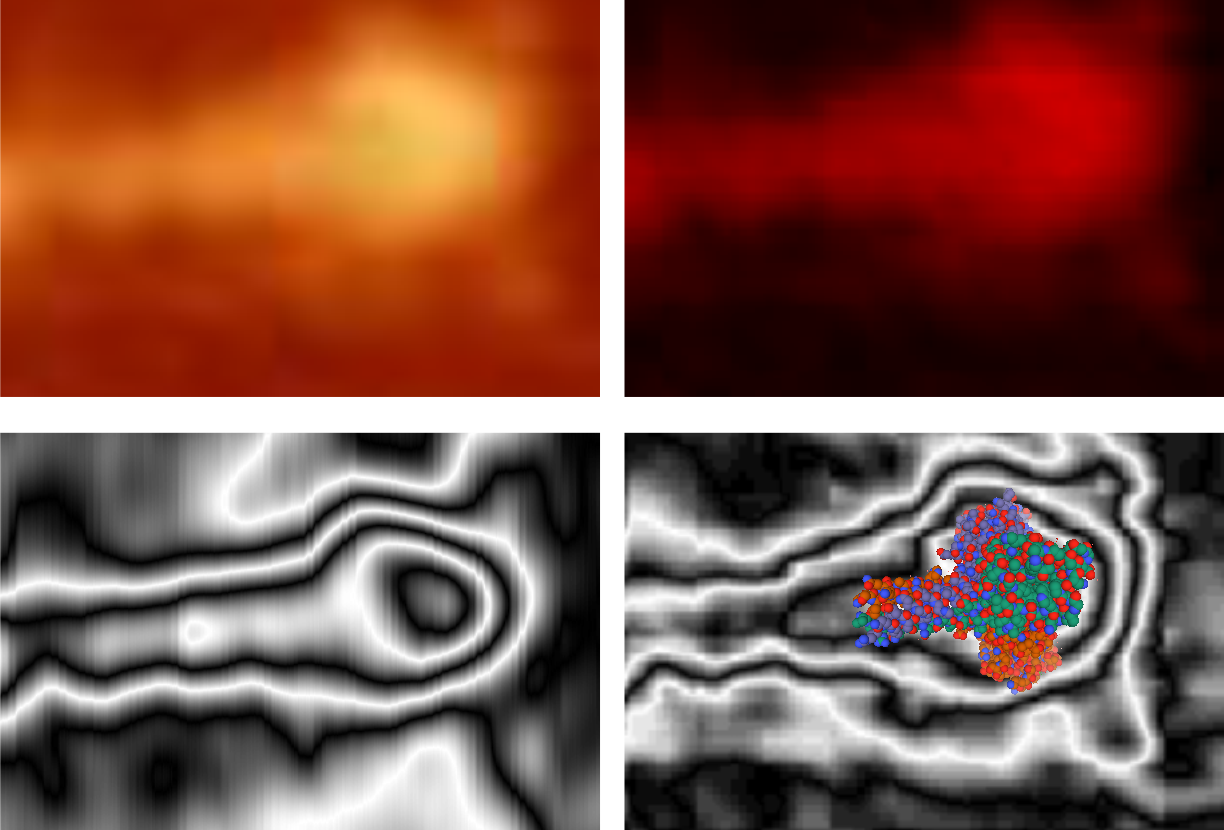
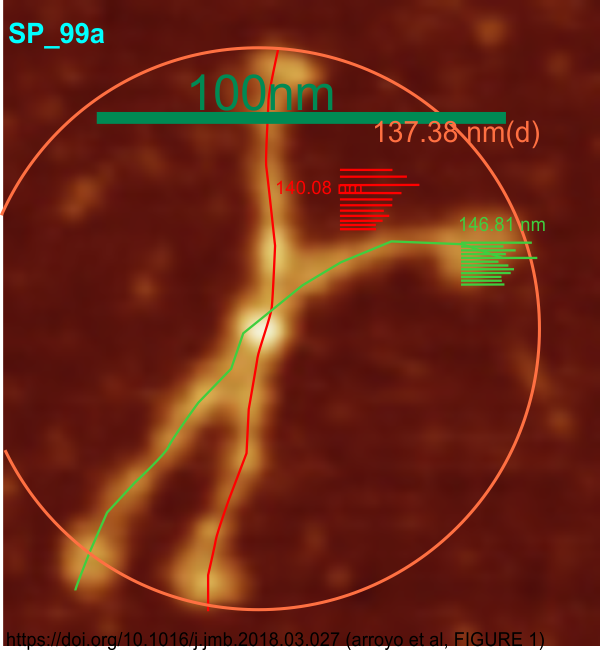
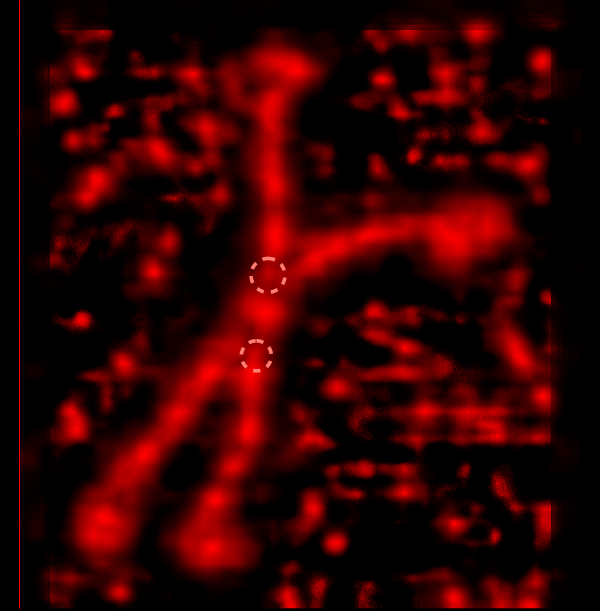
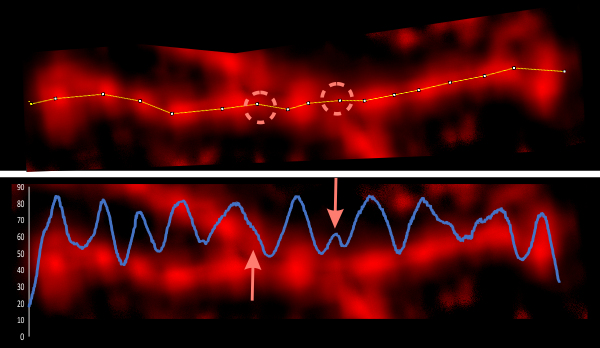
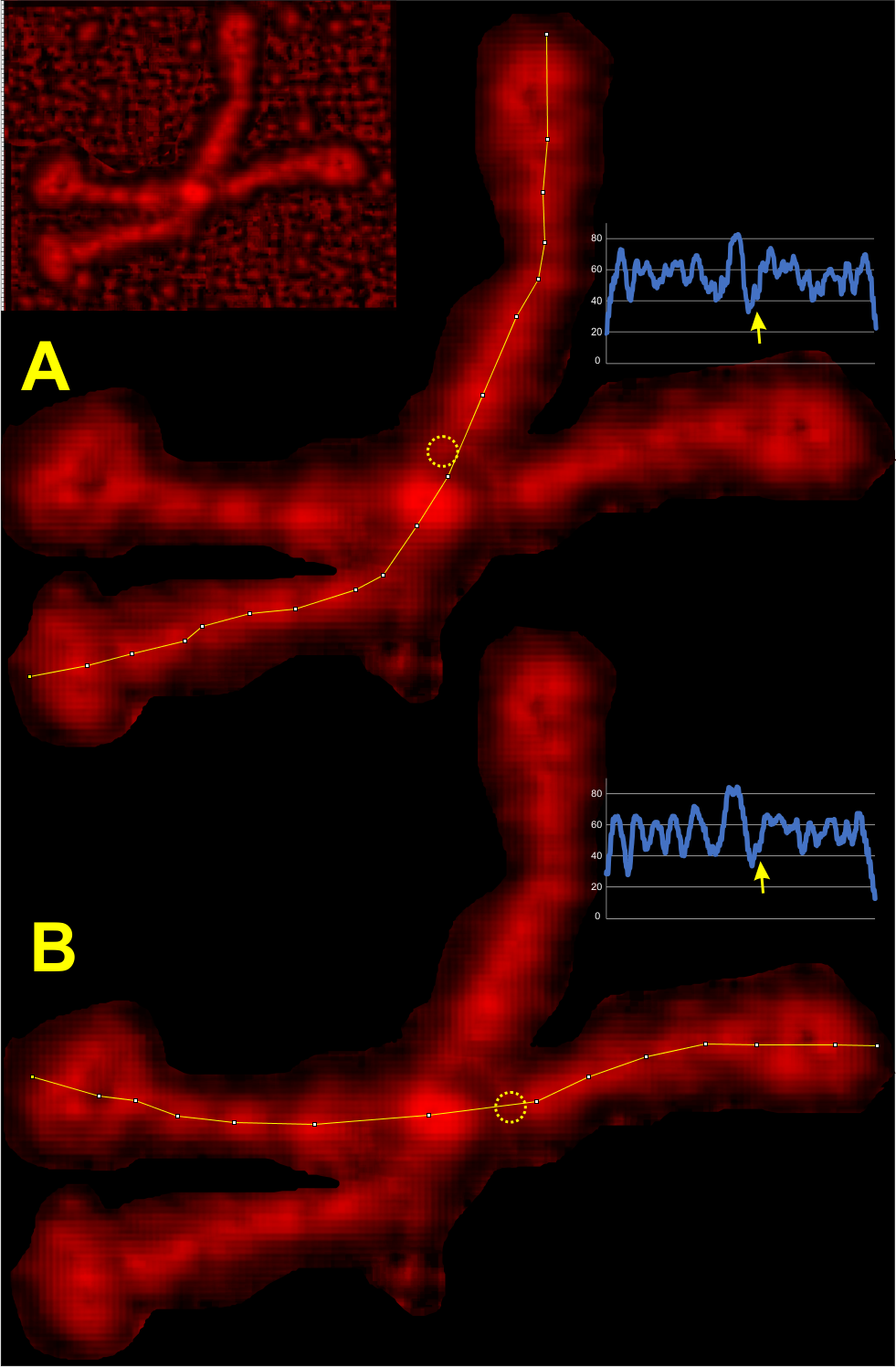
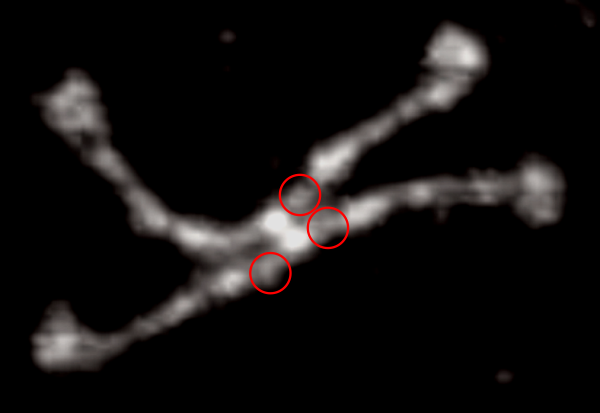
Some stereologic and morpometric methods I have used, invented for a particular purpose where no such method was previously described. It became a test of my ability to see variations in tissue whether methods I devised could ferret out what I “thought” I could see. If i saw a change, altered structure, greater or lesser incidence, odd shape or position after going through my slides or grids, I devised a way to acquire enough data points to decide if the change was significant. Rarely did anything I perceived as altered structure turn out to be chance…. — and could usually be sorted into an experimental group quite easily. Without going into examples (of which I could relate dozens) this is the dilemma for surfactant protein D. I have no doubt in my mind that there are between 4 and 6 “luminance” peaks between the N termini and the CRD for each of the trimeric arms.
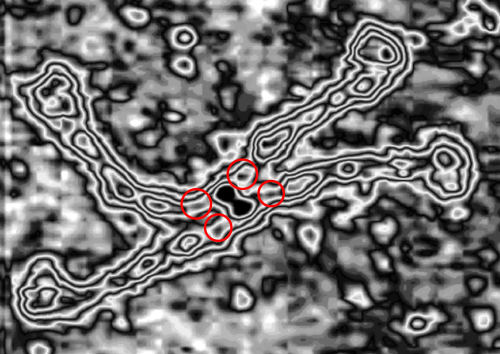
The dilemma is to figure out which data processing (as it is called in gwyddion) and called image processing by everyone else) tools can give me an accurate count of the peaks along the dodecamer arms. It is clearly reported by many SP-D researchers that the largest (height and width) peak is the N termini juncture in the center of a multimer, and the next largest peak (width height) are the 4 CRD at the end of each trimer.
Plots confirm what my eye’s see, but I am compelled to figure out how to add statistics to the observations. Using ImageJ (FIJI), CorelDRAW (which hands down is my favorite), Gwyddion, Photoshop, and now Matlab (which is way beyond comfortable for me) I have verified that many many ways of processing the images does not in any way change what I have seen. Yes, applying a mask or dialing down the brightness, upping the contrast, or applying any number variations to the RGB histograms can change the number, but most images from the beginning show a number of peaks and one can watch them disappear or increase in prominence…. at will. So when is image processing beneficial– what amount of removing background adjust for scan lines, correcting the data for scars and artifact is sufficient.
In retrospect, this project could have been just as valid if i had opened hundreds of images in photoshop and upped and lowered the contrast and brightness to a level i “liked” and hand counted the peaks… really. Then comes the sad answer, why am i doing this, to please some editor of some journal.
It would be ideal (nothing is ideal):
1- to have programs provide (and maybe they do, I just have not found it) documentation on which algorithms they use when inter-converting RGB or CMYK or grayscale or changing HSL or histograms.