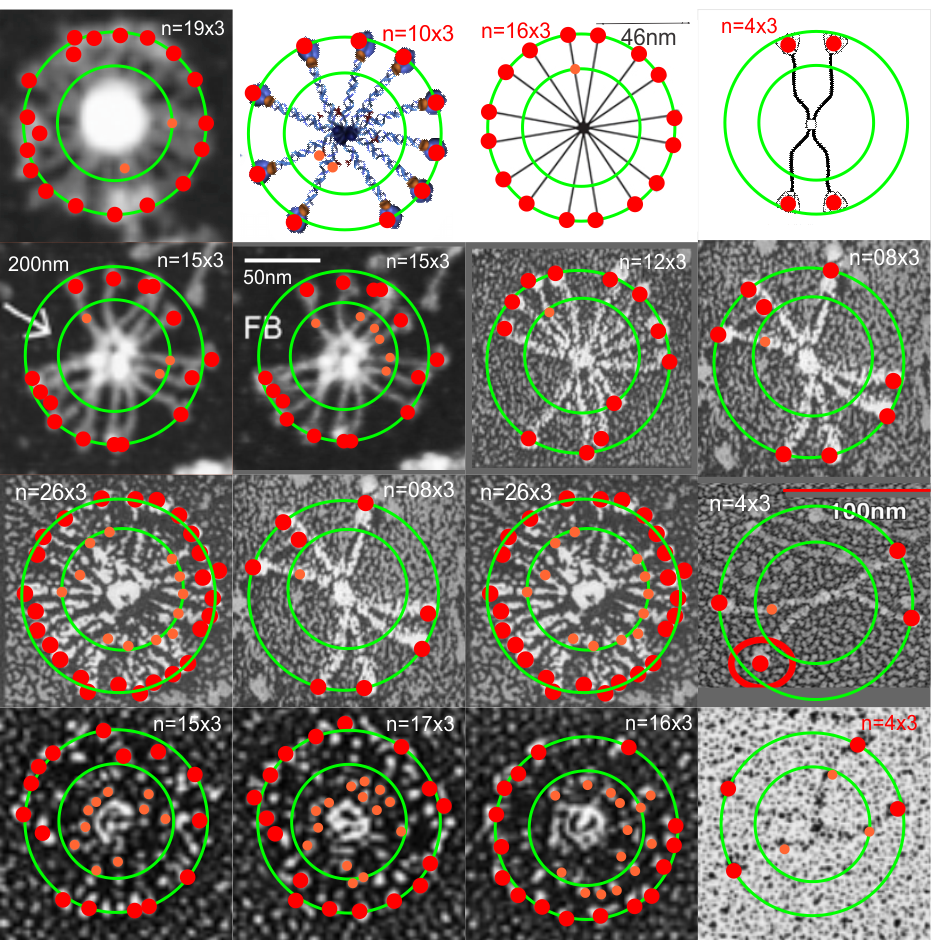
Arroyo et al have published what I think is a great paper using atomic force microscopy to look at many different multimers of surfactant protein D. There are a couple of things that I think they saw that might be what I also noticed looking at so many images of SP-D with various EM methods.
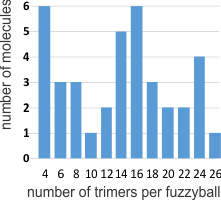
That is, there seems to be a blip or bulge in the SP-D structure close to the N terminal, and I marked that with some images in a previous post, then removed them for a summary page and the early video clip of SP-D. I think they are real, and also this publication indicates that it is the N glycosylation site…. and used some experimental manipulations to justify that view. Another issue is that the N-terminal sites of SP-D dont really often line up into a single area, they look sometimes like they are groups of separate arcs which have an angle of about 40 degrees (as measured on the micrograph below (the small angles) two angles were about 120 degrees. The center doesn’t look like a concentric area, rather a radially spaced loop-association.
This micrograph is from Arroyo et al, and it shows (as do many other images from their publication) what I am trying to indicate. Actually many of their fuzzyball micrographs show this the inner segmented ring rather than a single central density. I used their picture, and photoshopped it to eliminate background notations and debris, and other nearby molecules (you can see these in their original picture so you can compare) but I did not change the image of the molecule in any way. SO: one of the big issues is the curviness to the arms of the SP-D molecule (see my blue line in the image below), and another is the-off-center radially arranged N-terminal densities. These just make it unlikely that there is a “center” point where the N terminals come together in a single lump space. A third issue is that it seems like the angles between the hexamers (if in fact that is how they assemble into fuzzyballs… one hexamer at a time, or perhaps one dodecamer at a time, where the the angle is not that “acute” but is pretty broad, maybe on the order of 40 degrees. This is what the dodecamer shows most typically so there may be a reason to assume that in the fuzzyball that the 40 degree angle for the four arms (and a broader angle of 140 degrees between arms) is maintained. Blue ring (that I have added to image below) might be where other interactions in the fuzzyball structure bring the structure together in a sphere? or disc? at points of intersections of the “loops”. Other loops and centers around the blue ring are shown with white dotted lines. This particular fuzzyball one would prefer to see with even numbers of arms (they may be there)…. but apparently an arm or a couple of them got misplaced in the preparation process (LOL)… it is also my suspicion that there is a quantity “four” – that is, that the elements are divided into quadrants as reasonably convincingly shown in the micrograph below, and that works out to the 16 arms (an even number and multiple of 4) which pervades the surfactant protein D images that I have collected so far. White bar is 60nm according to Arroyo et al’s original image. Blue circle is about 40nm relative to their white bar marker.




 s
s
