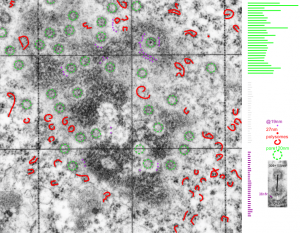
My quotes from an email to the PI of this project on 14CoS-/- mice treated with NTBC and withdrawn from NTBC. The electron microscopy on liver’s of these animals was really fascinating. Pan-apoptosis.”
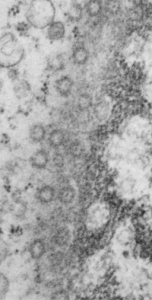
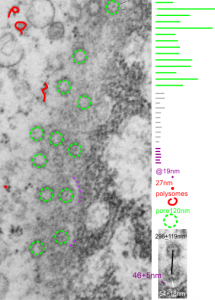
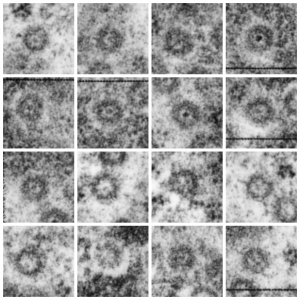
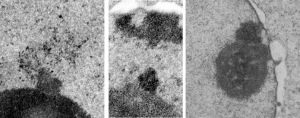
Take a look at the nucleolar structure, the dense body (likely denatured DNA and RNA) right next to remains of the a nucleolus and “notice” the regular pattern of striping (lower image, enlarged from square in right hand image above. Left image, earlier apoptosis with lace looking nucleolus with lots of fibrillar centers, and right, fibrillar center with dense RNA and/or DNA probably laddered.

“NTBC withdrawal in 14CoS/14CoS but not in ch/14CoS mice leads rapidly and progressively to apoptotic changes in hepatocyte nuclei. Early apoptotic changes were not associated with inflammation, however after 48 hrs some polymorphonuclear cells were present in the sinusoids. Isolated apoptosis need not induce an inflammatory response, however, when more three quarters of the hepatocytes are in some stage of apoptosis, the absence of inflammation is not a realistic expectation. The endpoint of both apoptosis and necrosis is in fact the same, cell death.” ”
“Mid-apoptosis is defined as hepatocytes where nuclei exhibit considerable peripherally condensed chromatin and unusually large nucleoli with very prominent fibrillar centers. Late-apoptosis is defined as cells where nuclei are pleomorphic in shape, have large clumps of chromatin condensed radially, an intervening nucleoplasm without texture, and show a disruption of nucleolar aarchitecture. Necrosis is defined as a swelling lysis of the entire cell, loss of membrane integrity, with debris everywhere. All histologic assessment was done using a blinded protocol. Percentages were calculated by dividing the number of apoptotic (or necrotic) cells by the total number of hepatocytes counted (>500 hepatocytes per animal), multiplied by 100.”
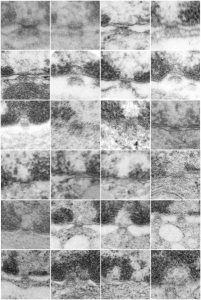
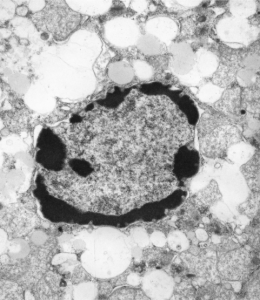
I am not sure how many or which of the countless images of hepatocyte nuclei ever made it into a manuscript but here is one image below where mitochondria have a very peculiar transformation.