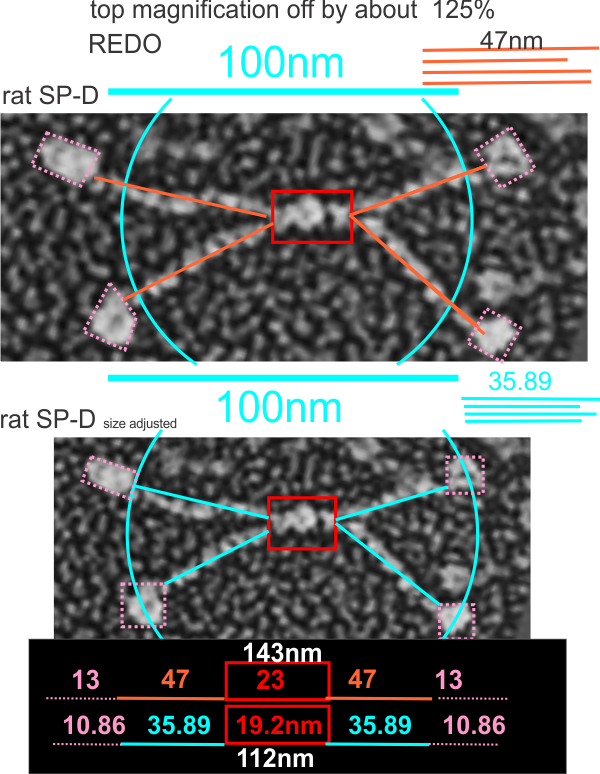
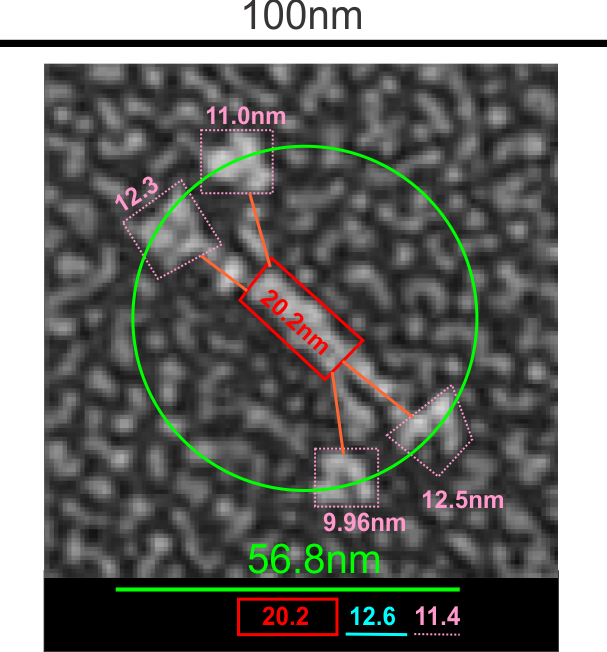
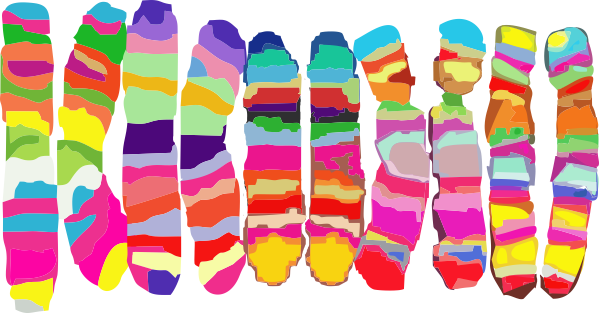
Here is where it become problematic when defining the size of anything as an absolute…. all these measures seen in publications need to be taken in a sense of wide variation in different reported sizes for the 100 nm micron marker. Case in point, from White et al, their 100 nm marker (my green bar underneath their own white nm marker) and my green circle (the same length as the diameter as my green bar marker) are shown without changing anything on their image….. this molecule is at least a 132% over what most individuals report as the alleged size of the 100nm SP-D dodecamer molecule. SO, if this rat SP-D dodecamer is that much bigger than the mini-SP-D which is claimed to be missing — “A rat mutant with deletion of two internal collagenous exons corresponding to exon 4 (C3) and exon 5 (C4) was generated by PCR overlap extension using full-length rat SP-D cDNA as primer”, how is one to know what the “real” size difference is.
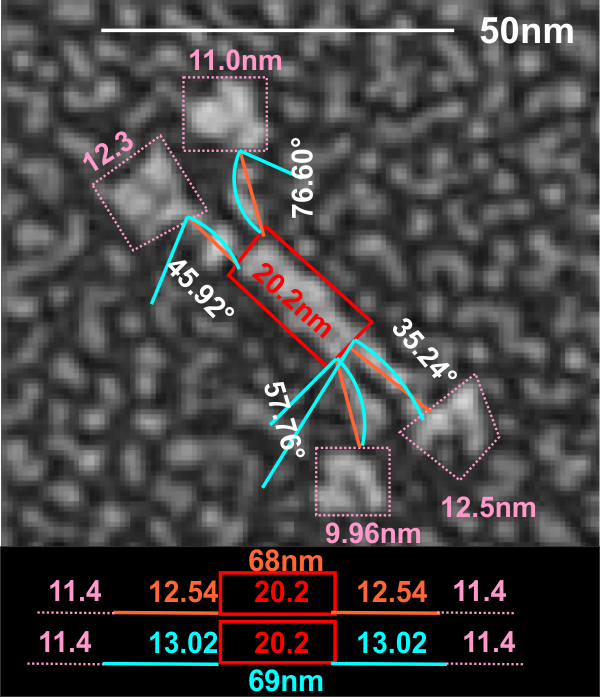
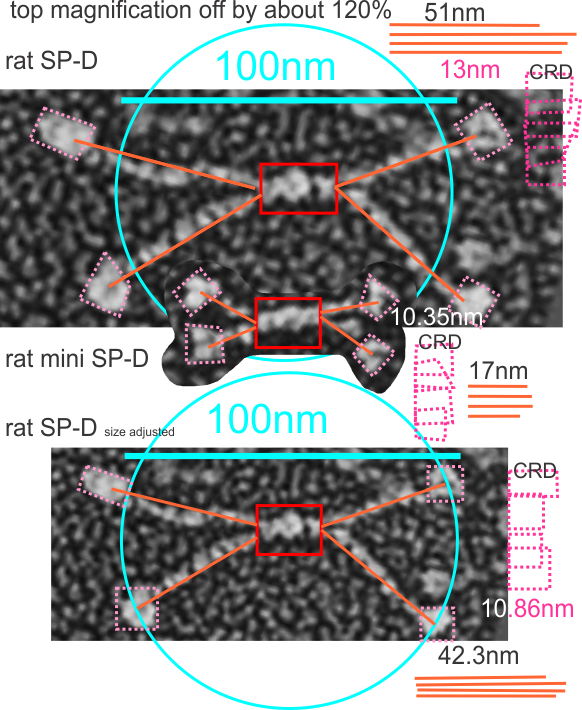
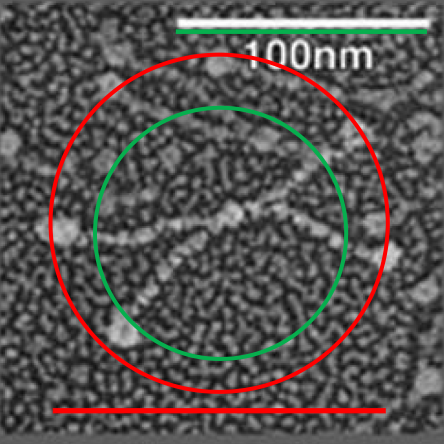 Here is an image using their own bar marker (green) and the diameter bar marker (peach), showing the relative decrease in arm length….. This is a 72% decrease from the rat SP-D in their own micrograph, and an additional decrease when using the “other researchers” estimates of 100 nm SP-D molecules.
Here is an image using their own bar marker (green) and the diameter bar marker (peach), showing the relative decrease in arm length….. This is a 72% decrease from the rat SP-D in their own micrograph, and an additional decrease when using the “other researchers” estimates of 100 nm SP-D molecules.
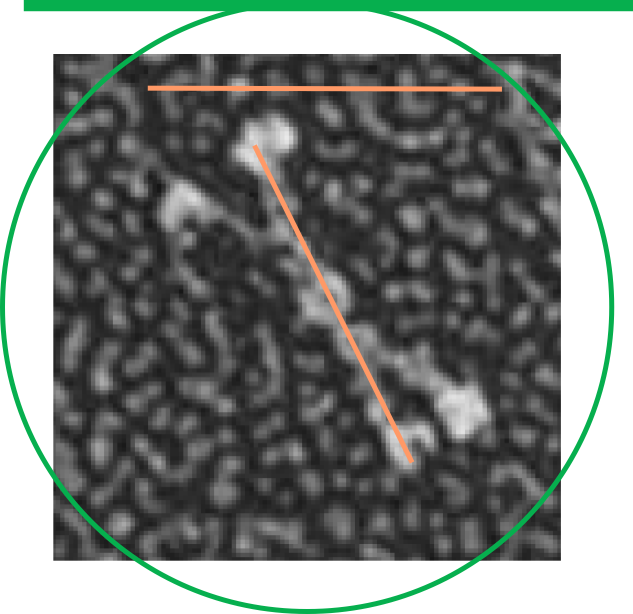
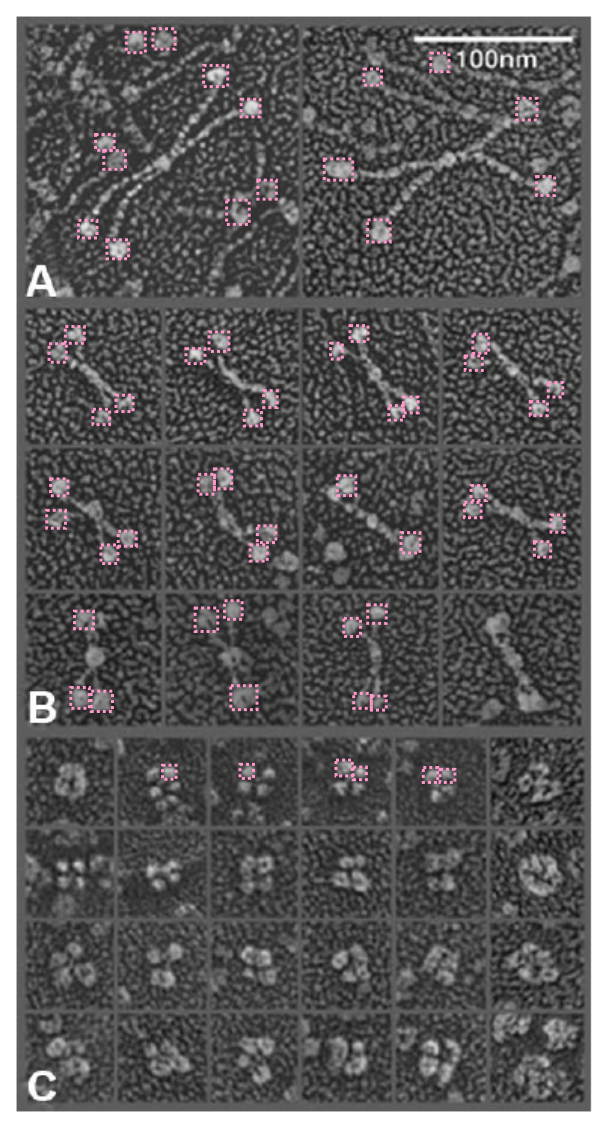
in trying to figure out the discrepancy in size of the molecules of SP-D in this publication i decided to do just a simple test: to take background images from the full SP-D and the SP-D dodecamers with the collagen portion deletions and see if they were anything similar in texture. So this also begs the question of differences in shadowing techniques on different grids… so that could be part of the problem but here clearly there is no similarity in the size of the shadow particles in the top (rat full length SP-D molecules) and the so called mini_SP-D dodecamers. This makes me think the full length SP-D is not the magnification reported.  From E. Crouch, et al 1994 “SP-D revealed a highly homogeneous population of molecules characterized by four or more relatively rigid-appearing subunits, non-randomly arrayed about a central core. The core was only poorly defined as the center of interaction for the N-termini of the subunits. The subunits seemed to emanate from opposite sides of the core in pairs closely apposed over a distance of 10 nm from the center of the core. The results for paired subunits presented here correspond very well to the structure of SP-D observed in electron micrographs. Site-directed mutagenesis of Cys-15 and Cys-20 has shown the necessity for N-terminal cysteine residues to stabilize oligomerization, but the cysteine residues are not necessary for trimer formation. However, other results provide strong evidence for the existence of at least one intra-subunit disulphide bond within the N-terminal region after limited peptic digestion.It is possible that apposing pairs of SP-D subunits are associated in an overlapping anti-parallel arrangement restricting the spatial distribution of the subunits around the central core. Another hypothesis is the association of paired subunits end to end. Two subunits are disulphide-bonded between and within subunits, whereas the two (or more) opposing pairs of subunits are associated through an undefined molecule. It has not been possible to exclude the possibility of the contribution of other molecules in the assembly of several SP-D subunits. It is also likely that cross-link formation between two or four subunits involves previously unpaired cysteine residue, or that a process of rearrangement converts a specific subset of intra-subunit disulfide bonds to inner subunit bonds.” this is good in a sense that when the centers of the fuzzyballs are examined, there is not an N-end to N-end “look” about the assembly.
From E. Crouch, et al 1994 “SP-D revealed a highly homogeneous population of molecules characterized by four or more relatively rigid-appearing subunits, non-randomly arrayed about a central core. The core was only poorly defined as the center of interaction for the N-termini of the subunits. The subunits seemed to emanate from opposite sides of the core in pairs closely apposed over a distance of 10 nm from the center of the core. The results for paired subunits presented here correspond very well to the structure of SP-D observed in electron micrographs. Site-directed mutagenesis of Cys-15 and Cys-20 has shown the necessity for N-terminal cysteine residues to stabilize oligomerization, but the cysteine residues are not necessary for trimer formation. However, other results provide strong evidence for the existence of at least one intra-subunit disulphide bond within the N-terminal region after limited peptic digestion.It is possible that apposing pairs of SP-D subunits are associated in an overlapping anti-parallel arrangement restricting the spatial distribution of the subunits around the central core. Another hypothesis is the association of paired subunits end to end. Two subunits are disulphide-bonded between and within subunits, whereas the two (or more) opposing pairs of subunits are associated through an undefined molecule. It has not been possible to exclude the possibility of the contribution of other molecules in the assembly of several SP-D subunits. It is also likely that cross-link formation between two or four subunits involves previously unpaired cysteine residue, or that a process of rearrangement converts a specific subset of intra-subunit disulfide bonds to inner subunit bonds.” this is good in a sense that when the centers of the fuzzyballs are examined, there is not an N-end to N-end “look” about the assembly.
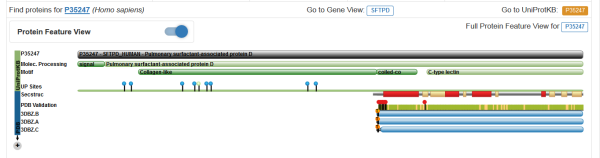
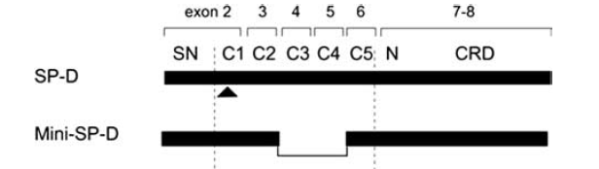
I am looking for the predicted length of the collagen-like domain with the C3 and C4 exons.
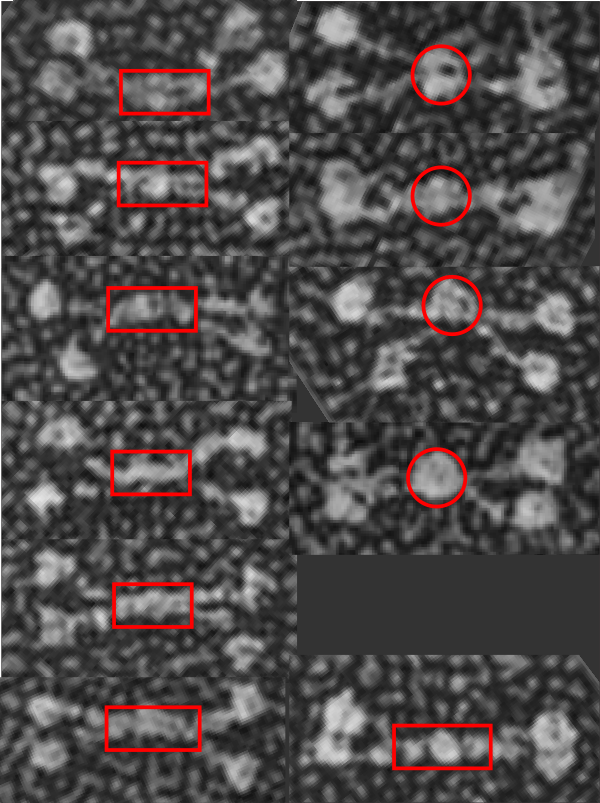 i think had i been doing this microscopy i might well have been advised to be absolutely precise in my measures of nm length. In retrospect in my own career…. i should also have been more careful.
i think had i been doing this microscopy i might well have been advised to be absolutely precise in my measures of nm length. In retrospect in my own career…. i should also have been more careful.