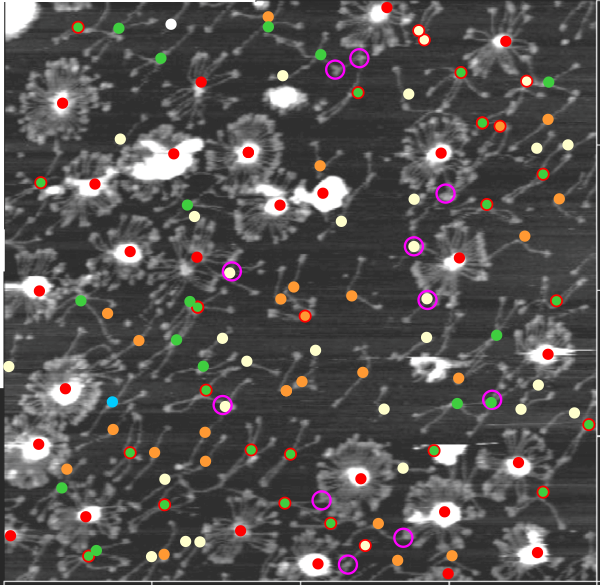
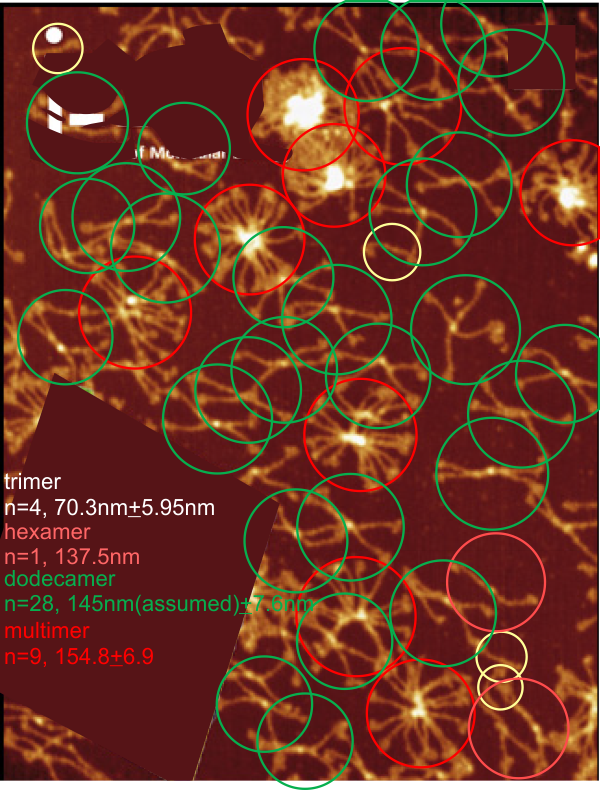
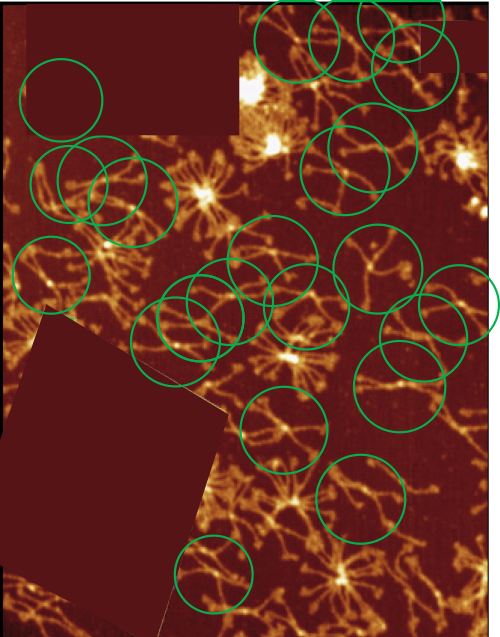
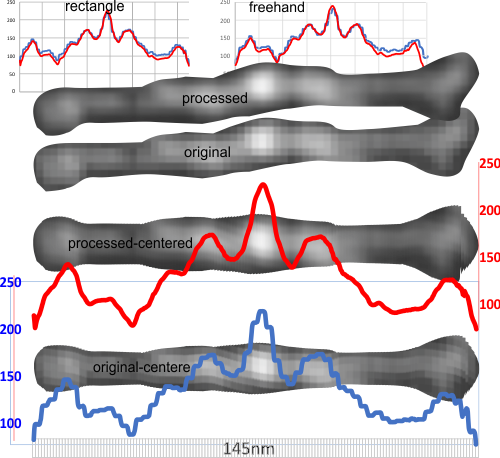
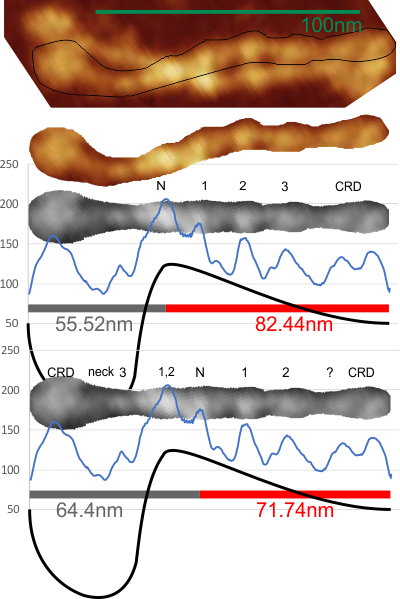
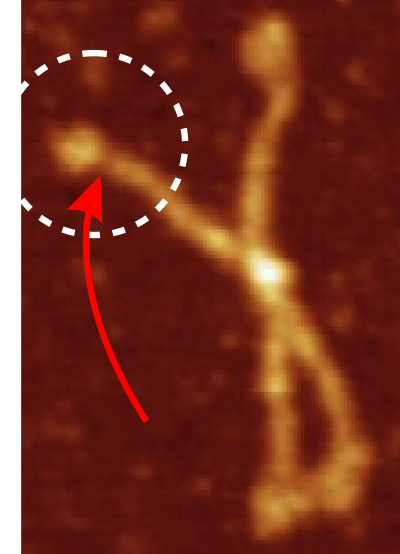
In many cases the CRD domain is in side to side proximity to other CRD, but sometimes the CRD is end to end or end to side with Ntermini as well. Some of these occurrences are denoted with a pink ring. Not all the molecules of SP-D in this image of Hartschorn et al (Respiratory Research) were marked, but the obvious trimers, one nonamer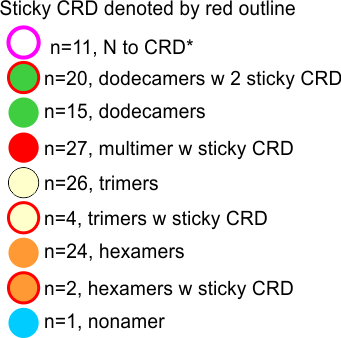 , lots of dodecamers and fuzzyballs were marked as to whether there were CRD which appeared to be in very close proximity – according to this legend.
, lots of dodecamers and fuzzyballs were marked as to whether there were CRD which appeared to be in very close proximity – according to this legend.
100% of the higher order (Mmore than 8 arms) SP-D multimers have sticky CRD (24/24); and about 57% of the dodecamers have sticky CRD (where two CRD come together in close proximity (and are bent and otherwise pulled with each other), 13% of the trimers have CRD touching some other structure (CRDs or Ntermini), and about 7% of hexamers touch something as well. Overall a third of the SP-D molecules have CRD that touch another CRD or an Ntermini –at least as it appears to me in this image.
Each of the 4 species of oligomer (except nonamers) are about 1/4 of the population. (thank you to Hartshorn et al)