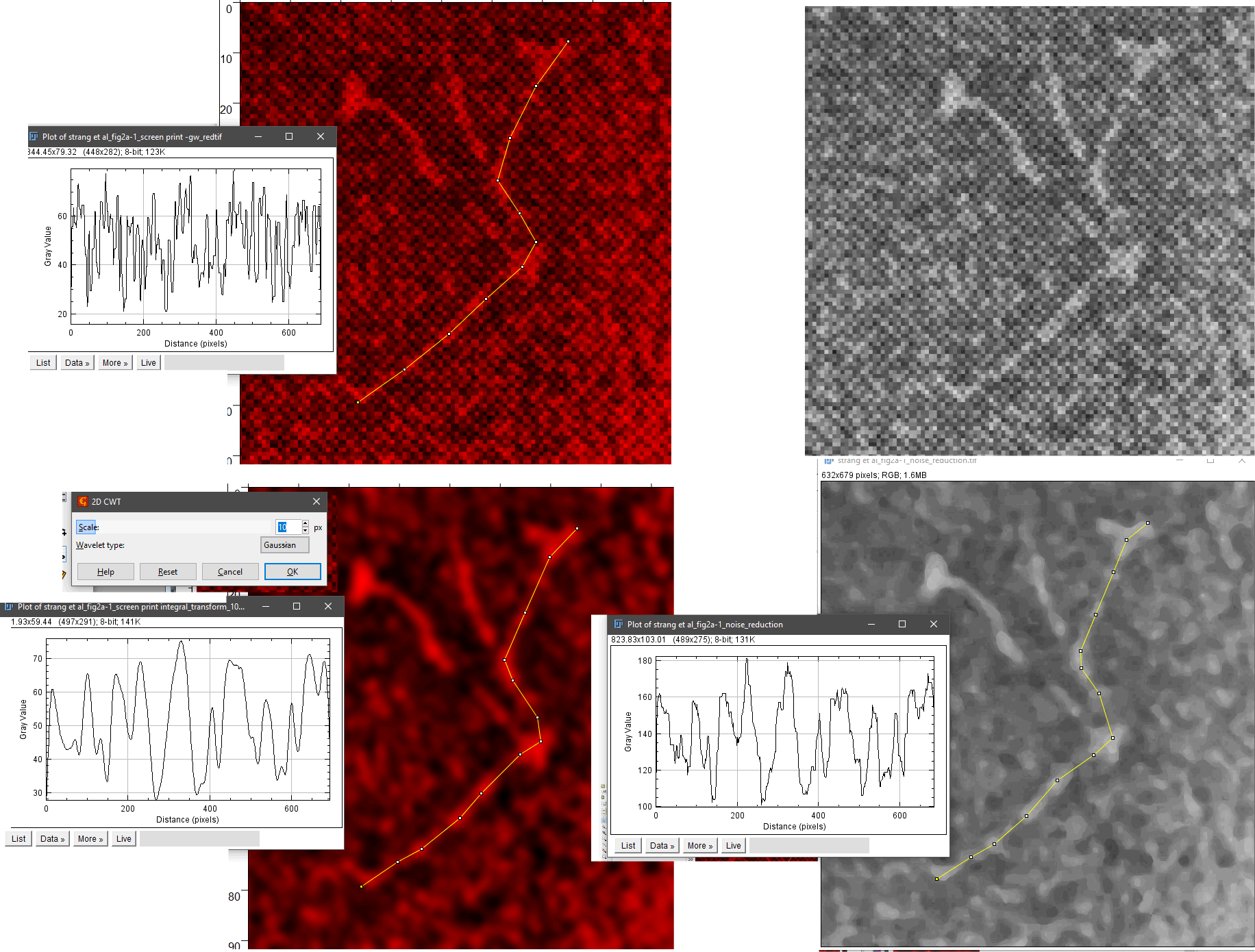
In trying to find the actual magnification for some early publications using negative staining and rotary shadowing to delineate the structure of surfactant protein D and also a closely-resembling member of the C type lectin group I ran into some images of bovine conglutinin (Strang et al, in the mid 1980s) that had images which were highly pixelated probably from the type of printing used in that era. I did a screen print of one of the negatively stained images (screen print of original here) and opened it in photoshop to play with different filters to reduce the noise. Gaussian blur and smart blur in photoshop did provide some improvement in the image but I decided to see what Gwyddion could do with the image in terms of processing out the noise.
Much to my surprise, a marvelous image emerged after applying “integral transform> 2D CWT with 10px gaussian blur. The imported image in gwyddion, and the transform are shown here. Upper right, screen print (on the computer keyboard) of a screen-printed (the technique for printing – likely and less expensive than offset printing) from the original article. (the pixels are not from the screen print of the image, as i did it at full screen capacity, they are from the screen used in the printing process). The image is poor quality at best. Image to the right of that is opened in Gwyddion as RGB, and image below on the left is a 2Dcontinuous wavelet transform of the image above, with gaussian blur set at 10px. The image on the bottom right is smart blur performed in photoshop. Each of these has had ImageJ used to mark a line through the molecule (by chance this bovine conglutinin molecule had just one arm that could be marked with a segmented line), and beside it is the plot generated in ImageJ. There is a tremendous difference in the information obtained from the 2D CWT processed image. Plots show the Ntermini junction, the CRD at either end and several plots along the collagen-like domain, so similar, yet so different from surfactant protein D. A great comparison. Nothing short of amazing.