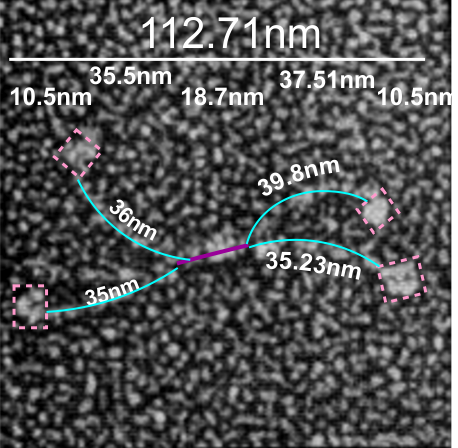
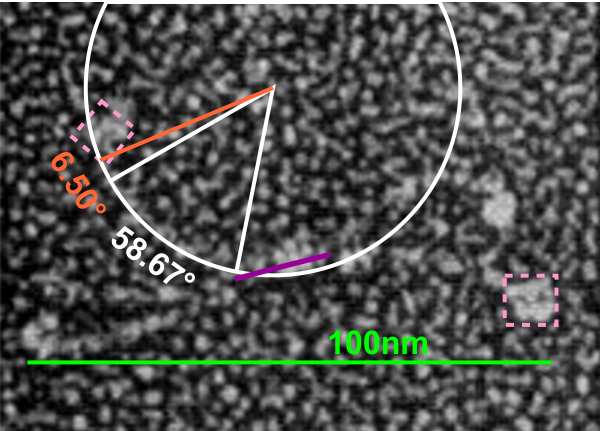
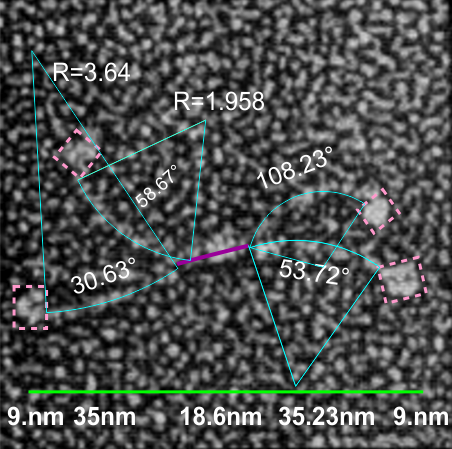
If i use the distance between two trimeric arms of SP-D seen in dodecamers as the off measured 100 nm width of the molecule, then measure the arc angle of each arm (arc angle length determined by [ 2pi R (central angle/ 360) ] this increases the total length (stretched as if a planimeter were used) of the separated collagen-like domain of the dodecamers at about 39nm for each half. (n=14 individual dodecamers with n=4 arms each), or the mean of the lengths of the two sides as separate individual measures (n=58 *one dodecamer had what looked to be five arms and was included).
The mean dimensions in nm of the N terminal domain, taken as one measure probably includes the N terminal as well as some small portion of the collagen-like domain, still both tethered together tightly (that is, the N terminal and a portion of collagen-like domain) and is about 19nm. The measurement spanned the area between bifurcation of the collagen-like segments on either end of the “joined” N terminal domains. (purple line)
The CRD measures 10.8nm whether determined by the mean of the length of the two sides, sorted by dodecamer (n=14 dodecamers, each with 4 CRD), or the mean of the lengths of the two sides all as individual measures (n=58 *one dodecamer had what looked to be five arms and CRD and was included).
A total dimension of about 119nm (the individual image shows a total dimension of 112.7nm – below). The very same diagram is shown above and below, one made before i decided to check how to measure the curvature as part of the length of the trimeric arms, the other after.
Here is the image posted earlier, where the arc length is NOT calculated, but just the straight line distance between where the trimeric arm diverges visibly from the center (Nterm plus whatever collagen-like sequences are there). When measured in parts, the total length of the molecul below measured at 107nm slightly above the 100nm which I constructed as the diameter between two CRD lying on a the circumference of a 100nm circle.
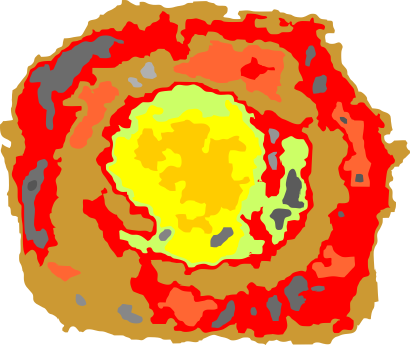
 What is pretty clear is that there are not that many, if any, accurate diagrams out there that have the proportions of CRD to thickness of the collagen like portion, or the curvature, or the length of the four segments of SP-D right.
What is pretty clear is that there are not that many, if any, accurate diagrams out there that have the proportions of CRD to thickness of the collagen like portion, or the curvature, or the length of the four segments of SP-D right.
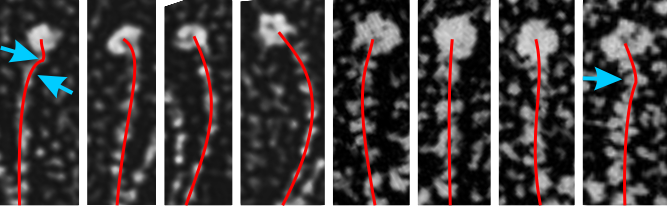
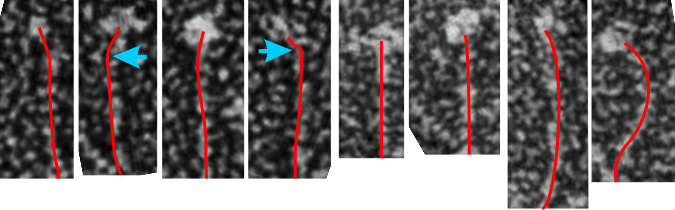
One thing for certain, is that shadowed and AFM images of SP-D show that the collagen portion is certainly NOT straight, as referred to in the chapter Alveolar Epithelium and Plumonary Surfactant, by Robert J. Mason MD and Leland G. Dobbs MD in the textbook Murray and Nadel’s Textbook of Respiratory Medicine, 8, 134-149.e5.
I could continue measuring all the dodecamer images i have collected using the arc length, central arc angle, N terminal (and whatever else) bound central segment, and size of the CRD but i don’t think adding those new data is going to change the outcome.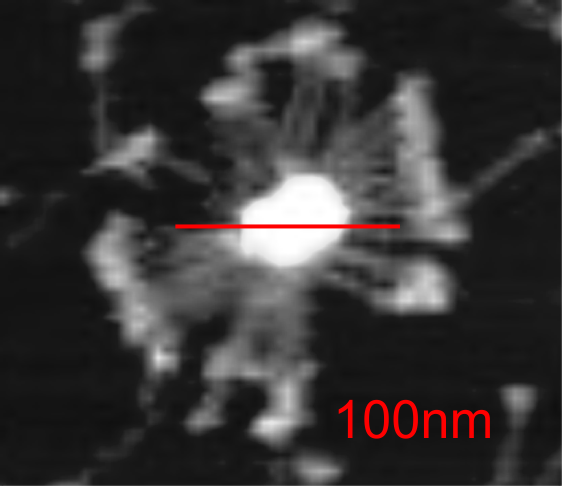
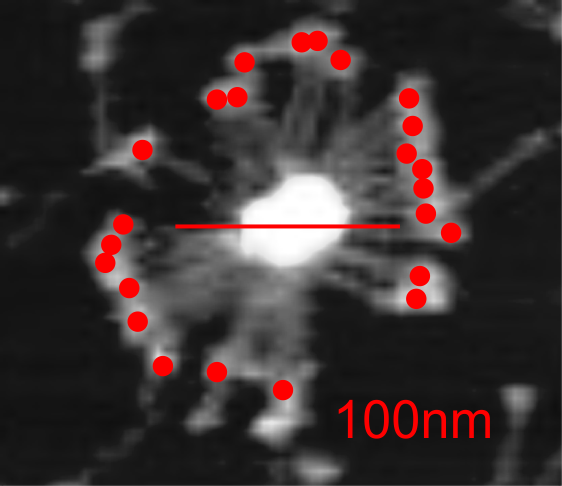
Here is a mutant surfactant protein D fuzzyball (from Kevan L Hartshorn, et al – with the mutation in the CRD so not likely affecting the overall dimensions much) which is greater than their own micron marker would designate as 100nm. This image was cut and pasted into photoshop from their figure, a dupllicate made, top layer increased in contrast, decreased in brightness, and layered at 50% transparency over their original in order to show the arms of the dodecamer and the CRD better. This is a mutant SP-D, according to them, with two substitutions in the CRD. (Recombinant human SP-D having a Met at amino acid 11 (and Ala at amino acid 160).
Counted the CRD just for the fun of it.