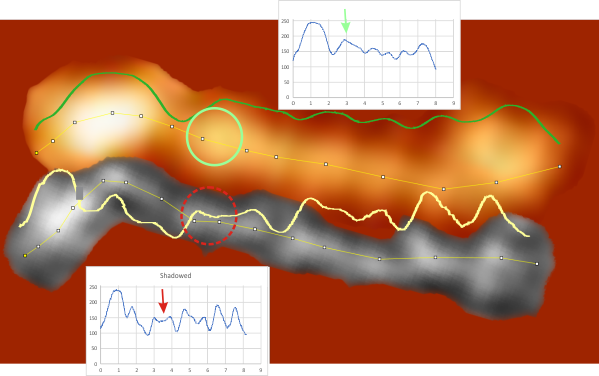
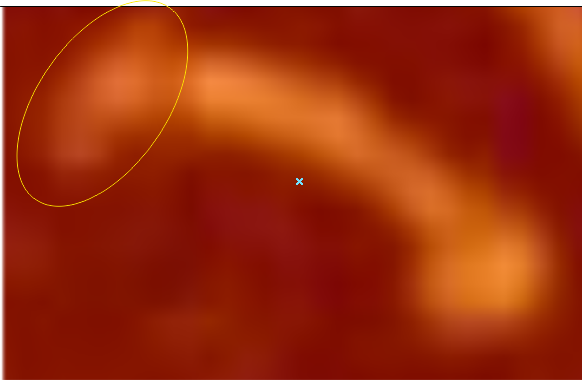
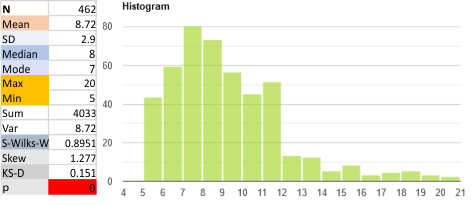
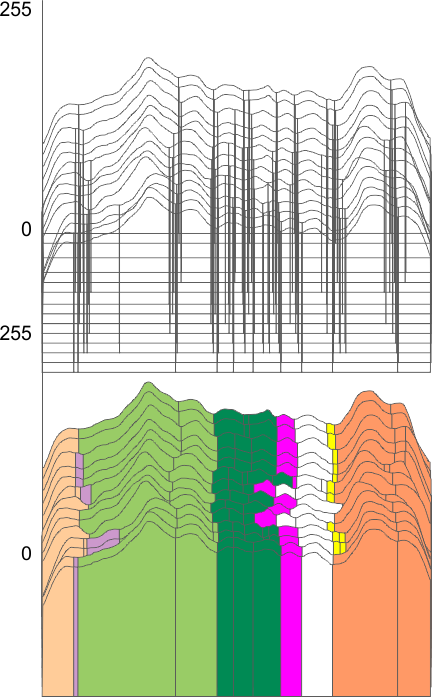
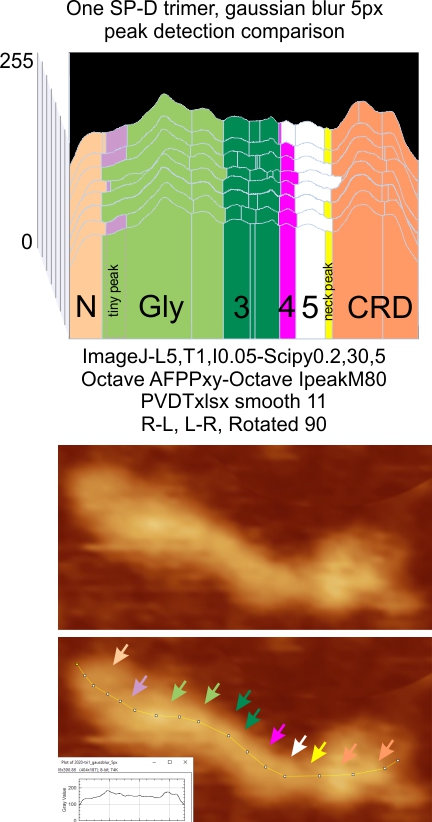
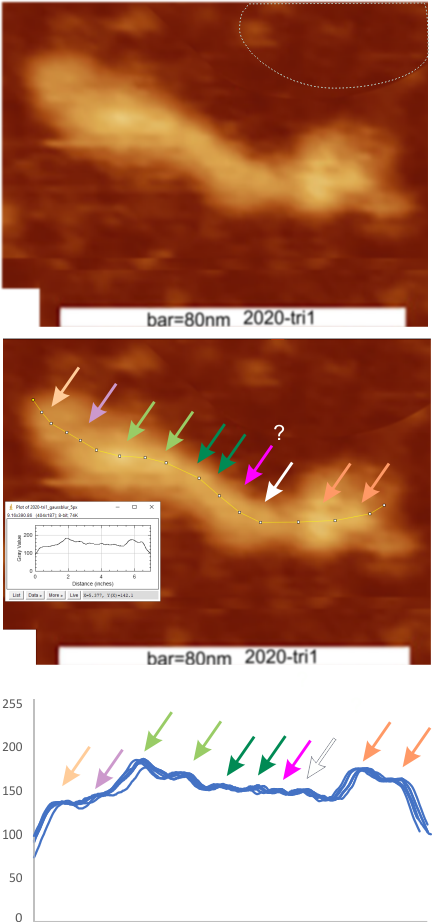
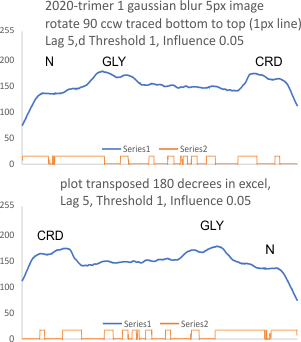
Parallel plots of a trimer of SP-D, AFM and shadowed TEM, took me about three seconds to pick two arms of SP-D, one from an AFM image one from a shadowed TEM image. No problem seeing similarities, the similarities in peaks were immediately apparent and similarities were pretty striking. Even though shadowed images have that typical lumpy background and it is a little difficult to ‘swallow’ the possibility of there being structural information in from both methodologies, the images confirm the number of peaks in a trimer of SP-D. i think the evidence which is just an observation here might be substantiated with more samples. The bottom line is that it is very clear that there are many more than the reported 3 peaks along the AFM and the shadowed images. The three known peaks ( N termini junction, glycosylation peak(s) and the CRD peaks).
Images were adjusted so that these “cropped out trimers” have the N terminus peak on the left, CRD peak(s) on the right and a grayscale plot was made to compare peak heights widths valleys, and of course peak number. Top image AFM, bottom shadowed TEM. In the two trimers below the N terminus peak is very bright since I cropped the trimers from dodecamers (where the N term peak is x4 where the four trimers are joined.
The difference between the top AFM image and the bottom that is immediately noticeable (and confirmed in other plots) is that the trimer in the AFM is glycosylated, so next to the N peak on the left, the bright peak is the glycosylation peak. The SP-D used for cropping out a trimer for the bottom part (shades of gray) of the shadowed image has not been shown to be glycosylated, so a glycosylation peak is not expected…… therefore that bright spot in the AFM denoting glycosylation is NOT that bright, more like a “low peak” in the gray shadow-cast image. Other peaks along the respective plots are very similar.
The biggest differences are 1) the tiny peak in the valley between the N term peak and glycosylation peak is prominent in the gray shadow-cast micrograph, 2) there is likely no glycosylation (or minimal) in the SP-D preparation that was shadowed (i have an AFM of that and will compare). 3) See the red circle for glycosylation peak on AFM image and not that bright peak where glycosylation would occur on the shadowed image (dotted line circle). There is a definite “foot” bend in the N term portion of the shadowed image, but also somewhat seen in the AFM image . The plots are very comperable 4) and the low brightness between the N and collagen-like domain is really prominent, and means something important in my opinion.

A new plot and set of images (3 this time) are below.
Since the biggest difference between these two arms, besides the methods of imaging, is the peak that is supposed to indicate glycosylation is different in all three. The top image below is definitely glycosylated (Arroyo et al, 2018) the AFM image at the bottom of the figure below is “possibly” partly glycosylated from the same authors. There is only ONE image of their deglycosylated SP-D dodecamers that I could find, from which the bottom SP-D trimer was cropped for comparison. The other three arms of that particular dodecamer labeled as deglycosylated SP-D had varying peak heights at the alleged glycosylation site. Whether one, two or three strands of a trimer are glycosylated seems to be an open question, and all the AFM images seem to indicate this since there is consistent peak brightness (height).
The particular trimeric arm of the deglycosylated SP-D molecule (so not a separate trimer, hence the high N term peak brightness), was selected not because of the glycosylation peak absence just because it had similar angles and would fit nicely one above the other. So the orientation was the primary selection bias, not the glycosylation peak height.
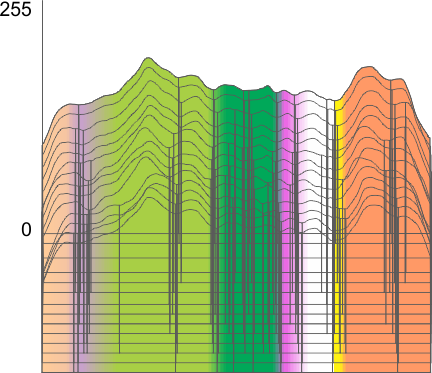
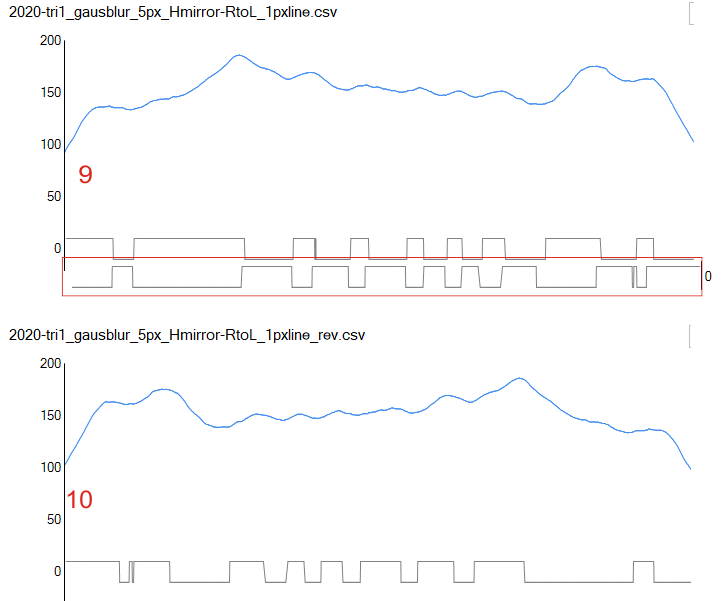
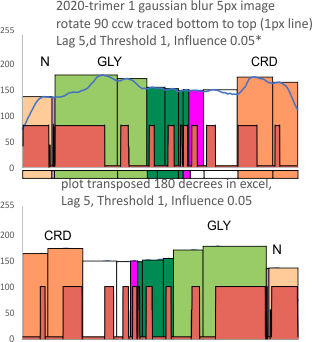
An assembled graphic (below) was prepared with molecules with an approximate shape, size and curvature to show three things: 1) the deepness of the valley between the N term and collagen like domain peaks, 2) the difference in the shadowed image (for which dodecamer the glycosylation state has NOT been determined, but it appears as if it is NOT glycosylated) and 3, the similarities of the peaks (excepting the variable glycosylation peak(s) of the three trimer sections of dodecamers with the two methods, and two separate preparations of SP-D. The grayscale plots on the right hand side of this image below shows the exact tracelines and the resulting plots for each trimer (obtained in ImageJ, exported to csv, plotted in excel). The exciting thing is that the peaks in the two prep methods (AFM and shadowed) are just really wonderfully similar.
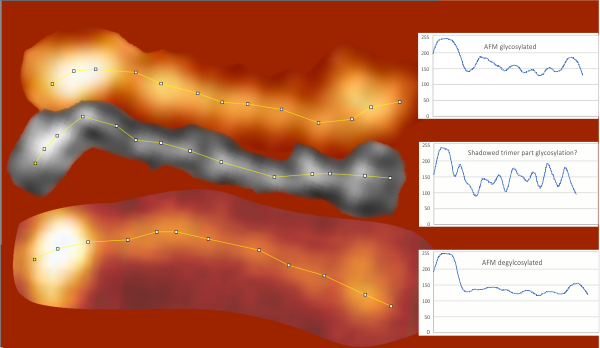 I had been skeptical of cropping out any SP-D arms to trace for grayscale levels in the shadowed image and just gave it a try… lest anyone accuse me of “doctoring” data…. no data have been changed, just the outlines. AND one really nice results is that the tiny peak that i have mentioned hundreds of times, that lies in the valley between the N terminus peak and the glycosylation peak is prominent in the shadowed image. If you look at the middle plot left hand side, you see the first tall Nterm peak, and in the shadowed image, a smaller peak on the downslope shoulder.
I had been skeptical of cropping out any SP-D arms to trace for grayscale levels in the shadowed image and just gave it a try… lest anyone accuse me of “doctoring” data…. no data have been changed, just the outlines. AND one really nice results is that the tiny peak that i have mentioned hundreds of times, that lies in the valley between the N terminus peak and the glycosylation peak is prominent in the shadowed image. If you look at the middle plot left hand side, you see the first tall Nterm peak, and in the shadowed image, a smaller peak on the downslope shoulder.