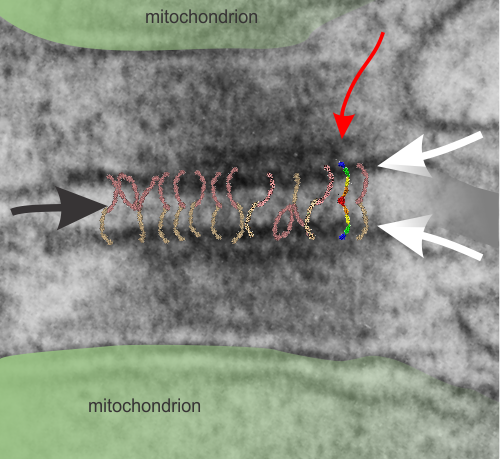
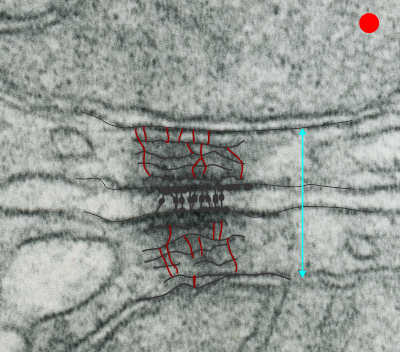
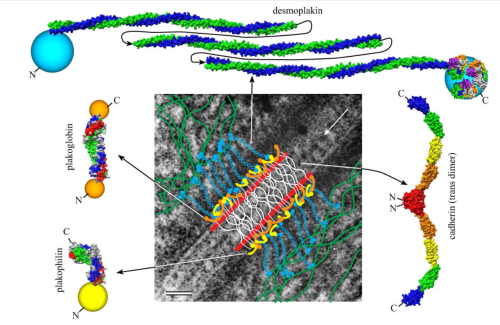
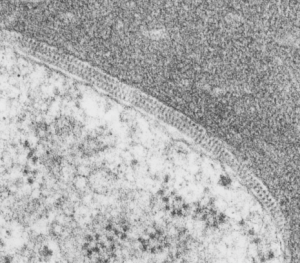
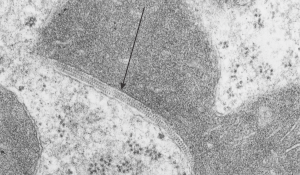
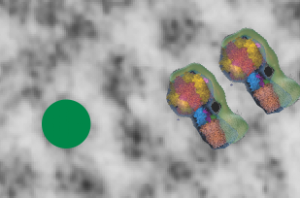
I found this particular electron micrograph which has two mitochondria and a desmosome in pretty good orientation to look at any periodicities or patterning in the substructure. It seemed to me that the central line of the desmosome, and the cadherin molecules which create it and the links to the plasmalemma (which have been modeled with molecular models) could fit best as a “sprint” type association. Those of you who are old enough to remember the plastic hair combs that looked like wishbones arranged in parallel that one could use to sort of tie up a pony tail, will recognize this flexible structure. I found, or should say “think i found” a similar type of density in the cadherin molecules in the center of this desmosome (diagrammed as that side by side “wish bone” array. Interestingly, cadherins were mentioned to one group to come in a dimer which would very easily form this springy wish bone structure…. and listed the “half wish bone” as one option as a molecular model of the desmosomal cadherins. One of those articles has a great 3D image which doesn’t look exactly like what i see, but is close found here – and another is this title Studer, Daniel & M Humbel, Bruno & Chiquet, Matthias. (2008). Electron microscopy of high pressure frozen samples: Bridging the gap between cellular ultrastructure and atomic resolution. Histochemistry and cell biology. 130. 877-89. 10.1007/s00418-008-0500-1.
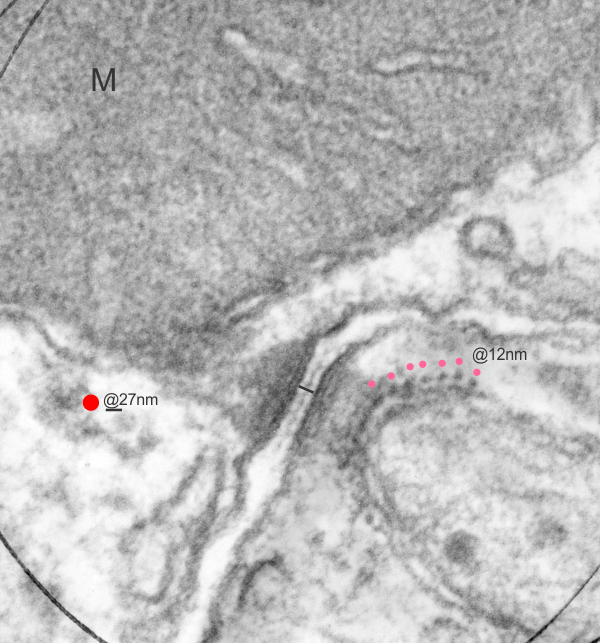
. I actually don’t like the idea of calling the arrangement “untangling desmosomal junctions with knots” as i think if they were knots….there would be no perfectly wonderful order that can easily be seen with ordinary transmission electron microscopy. It is also very appealing to have flexibility within these junctions as the wishbone (wish bone) alignment would afford…. that would be just really fun. 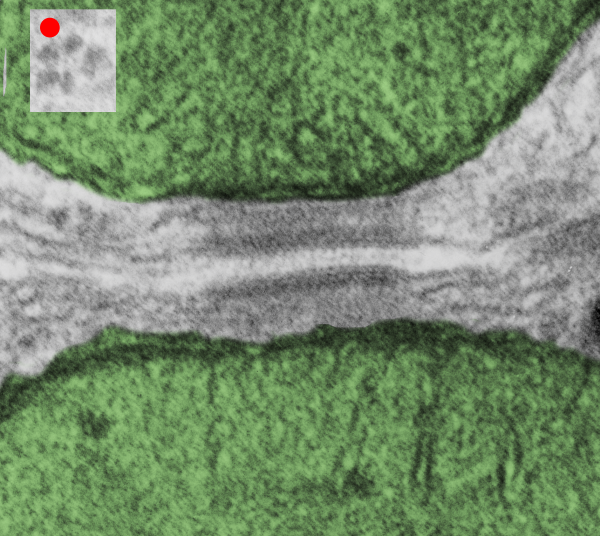 So here is the same electron micrograph as yesterdays post but the mitochondria are actually fuzzed green in a separate layer. Red dot=a ribosome, approximately 27 nm in diameter. The absolutely regular arrangement of cadherins is seen in the middle of the desmosome…. no knots, no tangles, no mess…. just regularly spaced. This micrograph is not retouched to emphasize anything…. it is just the way it looks on the negative. The plasmalemma of the two cells is not in exact cross section and so it is fuzzed. The span between densities both on the plasmalemmal sides and the center densities of this desmosome work out to be about 1/13-14nm. I have not put that together with any models yet and in the image below just sized the molecular structure according to the TEM, not using the actual size (just using the shape).
So here is the same electron micrograph as yesterdays post but the mitochondria are actually fuzzed green in a separate layer. Red dot=a ribosome, approximately 27 nm in diameter. The absolutely regular arrangement of cadherins is seen in the middle of the desmosome…. no knots, no tangles, no mess…. just regularly spaced. This micrograph is not retouched to emphasize anything…. it is just the way it looks on the negative. The plasmalemma of the two cells is not in exact cross section and so it is fuzzed. The span between densities both on the plasmalemmal sides and the center densities of this desmosome work out to be about 1/13-14nm. I have not put that together with any models yet and in the image below just sized the molecular structure according to the TEM, not using the actual size (just using the shape).
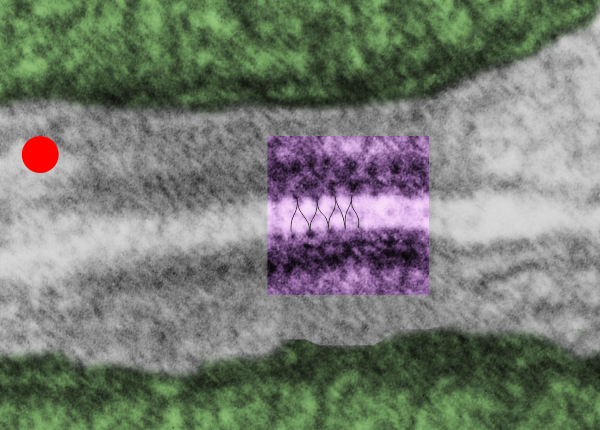
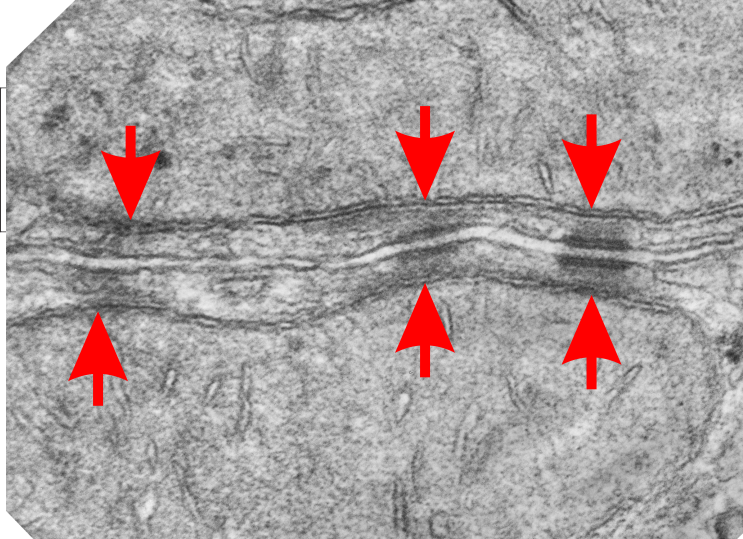
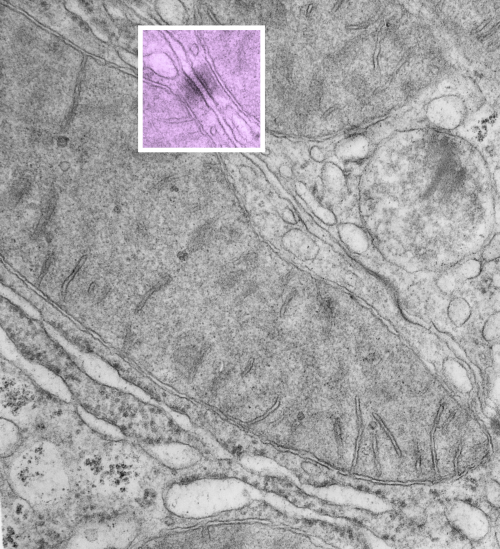
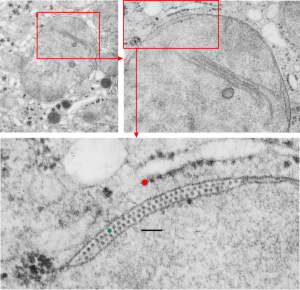
So in the second figure you can see an area that i enlarged, cut and pasted in photoshop, and enhanced in contrast and colored purple and ADDED what i think are the lines of the wishbone arrangements of the cadherins. Densities (increased in contrast here) work out to about 1 for every 13-14nm.
Into two enlargements (one the purple box above and another with the molecular model of a cadherin dimer, copied and mirrorred, fit really nicely into what might be a flexible, stretchy springy type portion of the desmosome. Just a thought…. ??
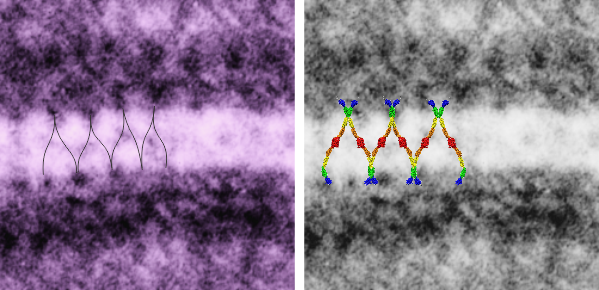
6110_5080_mouse_f_liver_36,200x_4x