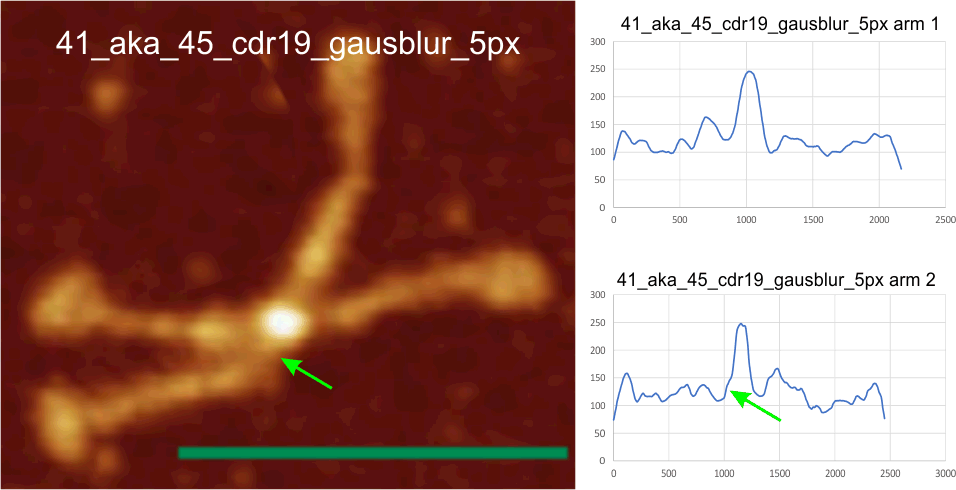
It is going to take a lot of convincing to persuade me that these four small peaks don’t exist: each I found in the trimers between the mostl unified peak of the N termini junction and the 4 glycosylation peaks on each of the trimers. It stands to reason that they will not all 4, always show up, in all AFM images, but then little does show up in every image, so thats not going to be a sufficient rebuttal.
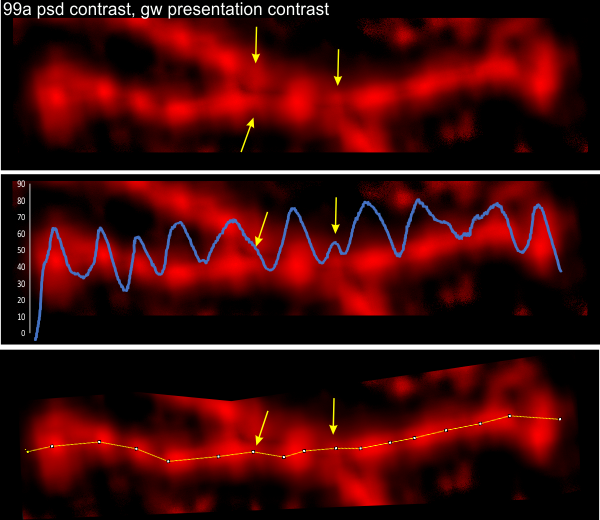
Here is an image from Arroyo et al, (which I call 99a). While three obvious tiny peaks can be seen in this image that has been processed (not signal processed, but image processed), the 4th one is there as a tiny bump on the downslope of the presumed glycosylation peak. Image on left shows tiny peaks pointed to by yellow in what i traced as “arm 1” and the green arrows show the tiny peaks in what I traced as “arm 2”. The image processing filters applied to this image was contrast enhancement in photoshop and limit-range in gwyddion. Relative peak heights in the plots to the right are skewed by the limit-range filter which eliminates all but a narrow range of brightness, but that does accentuate nicely the tiny peaks between the glycosylation peaks and the N termini junction (which on the plots lie above the arrows. Different dodecamers are seen for top-most image and the bottom 4 images. Image proessing does not seem to alter where and when the tiny peaks by N are found.