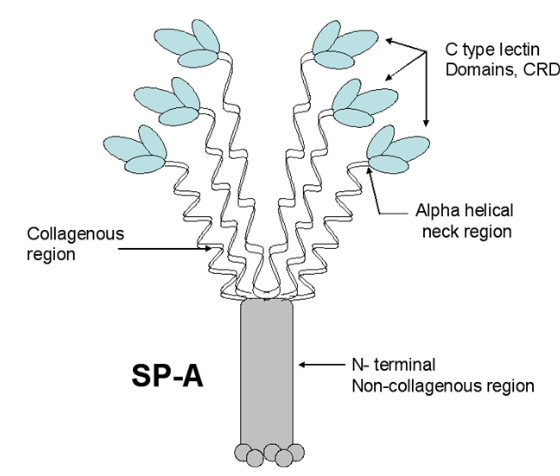
Remember playing “telephone” as a kid (also called “chinese whispers” — the title of which in and of itself denotes confusion)? Here is a link to wikihow if you never played it. The purpose is to show “in real life” what happens when information is casually passed from one to another (as in gossip… and i am calling the remake of scientific illustrations from previous publications as a similar phenomenon). The important truth is that information is easily corrupted. No more description is needed for the image below which is supposed to portray surfactant protein A. Clearly this artist/scientist team knew very little about SP-A or they were playing “telephone” with the specifics of how to draw the diagram.
What is sad to me is that this really bad diagram shows up in a place where many individuals might actually expect accuracy (a site called Global Science Books ) ? A link to the pdf that I found is HERE.
I will list what I think they missed, data from SFTPA1 online and the literature.
- The ratio of AA for each of the 4 domains seems to be wrong, as the N terminus is very short, the collagen like portion is pretty long (blue), coiled coil neck domain (black) is not too long, the CRD (red) is quite long. The relative distances of these four domains in this diagram are quite far off, in spite of the fact that many much better diagrams are available for free. Signal peptide is 1-20.
- Where did the third strand of the collagen-like domain go? It looks to me like the two twined black lines in their diagram are just that “2” not “3” as repeatedly mentioned in texts (every text i have read in fact) and why would it be called a trimer if not three, and how in the world does the third CRD domain hook up when there is no line in the collagen like domain to which to attach it?
- There is virtually no distinction between the winding of the collagen-like domain and the coiled coil neck… an arrow points to the transition, but there is not definition, and as well, there are only 2 of the 3 strands in the neck domains present.
- It is difficult to interpret the trunk like portion of this diagram, and how it might relate to the interactions between the collagen like domains, of the 6 trimers. It might be noted that in their diagram (not shown here) lying right next to this SP-A diagram shows the dimer of SP-B in the same two lines as the trimer of SP-A? Why?
- What in the world are the “balls” circles at the N terminus? Is this a depiction of N termini binding sites?
- Why are the three supposedly nearly (if not absolutely) identical CRD pictured in different sizes?. If it is an attempt at forshortening in graphic design if fails miserably.
OK, this is enough, you get the point.