I found a nice review article the title is “Intermediate Filaments as Organizers of Cellular Space: How They Affect Mitochondrial Structure and Function by Nicole Schwarz * and Rudolf E. Leube” free online, about intermediate filaments and mitochondria, which is part of my reference list as I am trying to figure out where there are species differences in the ultrastructure of desmosomal-intermediate filament-mitochondrial tethers. So this is a nice diagram but i think it is too primitive, or perhaps too limited, or maybe just has not been taken to the next step.
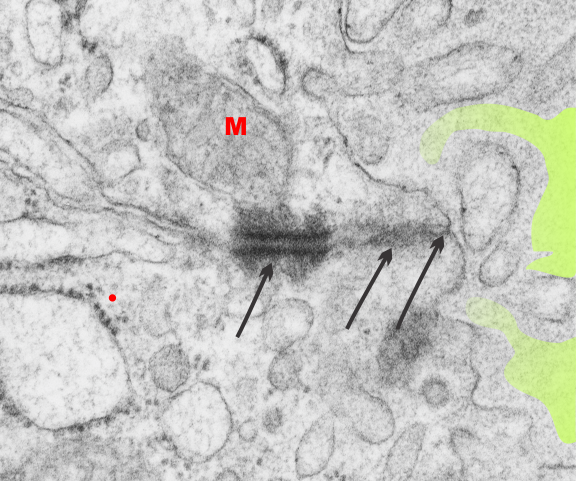
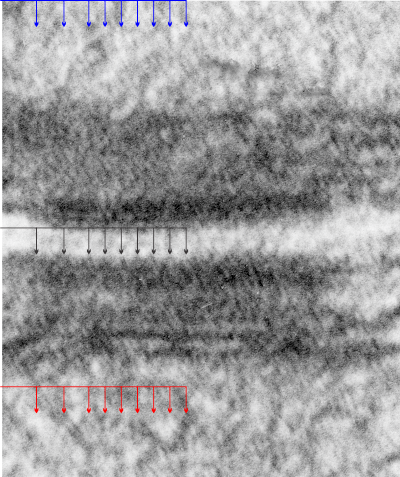
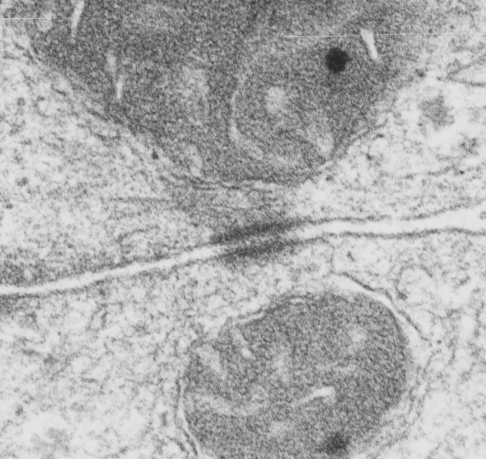
This image is their Figure 1 which shows three possible intermediate filament – mitochondrial arrangements. My exception to it is the following : there are areas around each mitochondria where all these types of bonding-binding-exchange take place, at least it seems to me. For instance, the binding of intermediate filaments to the outer mitochondrial membrane (OMM) probably doesn’t occur (at least in hepatocytes) in a complete manner surrounding the entirety of the outer mitochondria membrane, but rather definitely is “focal” as it looks when it is tethered to another cytoplasmic organelle such as the outer edges of the nuclear pore, or the inner plaque of the desmosome. There is a definite change in the structure of the outer mitochondrial membrane, gross enough that it is seen with TEM, and looks like a thickening of the OMM and a “flattening” out of that area which is engaged with intermediate filaments. There is also a shape change to the mitochondria when they are bound (i prefer the word tethered) to the filaments. (so still talking hepatocytes here). Intermediate filaments positioned remotely from the OMM and called “signaling” is something that I don’t believe would be seen physically. As for confinement, there are certainly areas of confinement of organelles, and they might be called “no fly” zones, I also need to find an example where the mitochondria are almost repelled by intermediate filaments.
mitochondrial arrangements. My exception to it is the following : there are areas around each mitochondria where all these types of bonding-binding-exchange take place, at least it seems to me. For instance, the binding of intermediate filaments to the outer mitochondrial membrane (OMM) probably doesn’t occur (at least in hepatocytes) in a complete manner surrounding the entirety of the outer mitochondria membrane, but rather definitely is “focal” as it looks when it is tethered to another cytoplasmic organelle such as the outer edges of the nuclear pore, or the inner plaque of the desmosome. There is a definite change in the structure of the outer mitochondrial membrane, gross enough that it is seen with TEM, and looks like a thickening of the OMM and a “flattening” out of that area which is engaged with intermediate filaments. There is also a shape change to the mitochondria when they are bound (i prefer the word tethered) to the filaments. (so still talking hepatocytes here). Intermediate filaments positioned remotely from the OMM and called “signaling” is something that I don’t believe would be seen physically. As for confinement, there are certainly areas of confinement of organelles, and they might be called “no fly” zones, I also need to find an example where the mitochondria are almost repelled by intermediate filaments.
In a couple of mouse models i have looked at, intermediate filaments themselves can be repelled (or it looks that way), of which one is the central confinement of intermediate filaments in small intestine microvilli where the gastric HKatpase is missing). There are a dozen, at least, intermediate filament x OMM sites that can be seen hepatocytes where there is obvious “tethering”, and i bet signaling as well. So these categories might have a lot of crossover.
I am thinking that in order to signal, the intermediate filaments need to be physically closer to the mitochondria than this diagram projects…. I wonder how that could be determined with TEM.