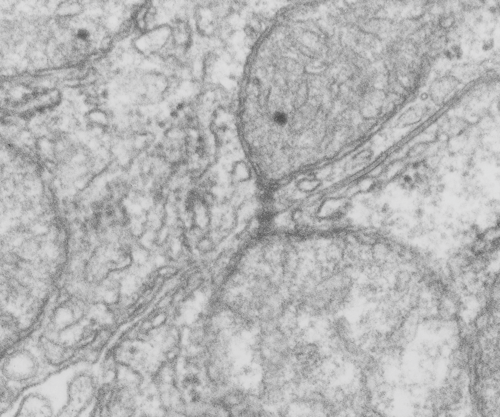
Two tiny mitochondria associations with a tangential section of a desmosome. Hepatocyte. 5893_2222. Not a particularly stunning micrograph, but sure documentation that mitochondria associate with desmosomes in hampster liver parenchymal cells.
Monthly Archives: December 2017
Who could guess that desmosomes know hebrew?
Was looking at this model for the cadherins of a desmosome and I just happened to see “shin” there in reds and oranges. I love it when nature intersects civilization in an unexpected manner…. so fun, ha ha. Just out of deference to the authors of this paper on desmosomes, 3D and electron microscopy I will list their reference in case you want to actually see the original. Al-Amoudi, et al., Nature05994 (2007) 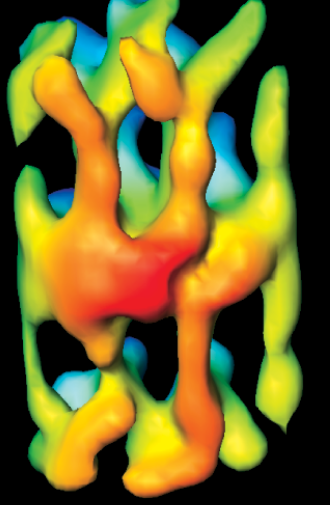
You’ve got to be kidding me!!
 WHo does this. Nutrogena named their baby product… “pure and free”. Have you read the ingregients. I hope i spelled them all correctly. If you don’t know some of these, google them. Some of these names i
WHo does this. Nutrogena named their baby product… “pure and free”. Have you read the ingregients. I hope i spelled them all correctly. If you don’t know some of these, google them. Some of these names i
know because i used them to embed tissue for electron microscopy. Not for anyone’s skin. I listed the 4 “OK” ingredients at the end….. these are not even the active ingredients, which include titanium dioxide and zinc oxide. 36 chemicals I don’t want to use on my skin.
butyloctyl salicylate
bees wax
styrene/acrylates copolymer
silica
butylene glycol
polyethylene glycol 8
glyceryl stearate
polyethylene glycol 100 stearate
cetyl dimethicone
alumina
benzyl alcohol
dimethicone
polyhydroxy stearic acid
arachadyl alcohol
methicone
hydroxyethyl acrylate/sodium
acryloyldimethyl taurate copolymer
xantan gum
isohexadecane
behenyl alcohol
trisiloxane
trimethy siloxysilicate
ethylhexylglycerin
disodium EDTA
arachidyl glucoside
dipotassium glycyrrhisate
bisabolol
triethyxycaprylylsilane
betahydroxy toluene
polysorbate 60
stearic acid
methylisothiazolinone
polyaminopropyl biguanid
polymethyl methacrylate
glycerine
retinyl palmitate
avena sativa kernal extract
:
tocopheral acetate
ascorbic acid
pantothenic acid
water
More on desmosome shape
“Desmosomes are a complex assembly of protein molecules that form at the cell surface and mediate cell–cell adhesion. Much is known about the composition of desmosomes and there is an established consensus for the location of and interactions between constituent proteins within the assembly. Furthermore, X-ray crystallography has determined atomic structures of isolated domains from several constituent proteins. Nevertheless, there is a lack of understanding about the architecture of the intact assembly and the physical principles behind the adhesive strength of desmosomes therefore remain vague. We have used electron tomography to address this problem. In previous work, we investigated the in situ structure of desmosomes from newborn mouse skin preserved by freeze-substitution and imaged in resin-embedded thin sections. In our present work, we have isolated desmosomes from cow snout and imaged them in the frozen unstained state. Although not definitive, the resulting images provide support for the irregular groupings of cadherin molecules seen previously in mouse skin.” ABSTRACT from Gethin Rh. Owen, Devrim Acehan, K.D. Derr, William J. Rice, David L. Stokes . Some things might be right, others not…. the last idea that cadherin molecules are grouped irregularly i don’t like. (This article i wont read as it is pay-per-view, which really galls me particularly because most research is paid for by tax-dollars and should be free).
” I found a dimension for the width “wide intercellular space (~240 A)” at 24nm and a measurement of the intermediate filament at 8nm (i presume diameter). The 22-24nm (could be less in some instances more like 15nm) as the width of the desmosomal intercellular space (which includes the densities on the outer leaflet of the plasmalemma from both cells and the central dense line of the desmosome. That measurement looks about right. From this I estimate the width of the electron lucent annulus of the desmosome (which is much more evident when there is a tight junction on either side of the desmosome) to be dependent upon where the section sectioned the spot desmosome, and whether the cut was equatorial or tangent to the spot itself. On one micrograph (stub tail monkey hepatocyte) in which one area of the desmosome is close to a tight junction, shows this lucent area (the outer annulus) to be about 35nm in its radius – that dimension applies to the edge of the central dense line extending to the tight junction. The width of that particular desmosome (where the central line was apparent) was @265nm. This makes the ratio of the dense desmosomal central line density distance to lucent annulus distance in cross section about 3.5%.
That lucent area may not be a uniform annulus, but if it were even, this assumes pretty much a “round” desmosomal spot weld. In measuring the radius of the annulus (just in the part ring part) I understand that the central dense line of the desmosome will appear in any view with the spot perpendicular to the plane of section, and this doesn’t diminish the fact that closer cuts to the outer edge of the desmosome will have a greater annulus to spot ratio. Fuzzing of the central dense line in the desmosome occurs when the angle of the section is not perpendicular to the spot, and can happen either at an equatorial section or anywere past that equator. So. any question I can ask, someone has an answer to…. so here is the definition of calculating the area of an annulus.. ha ha “An annulus (Latin word for ring) is a two-dimensional region of space bounded by two concentric circles. The area of an annulus is the difference in the areas of the larger circle of radius R, and the smaller circle of radius r. Mean radius (ρ) is the average of the exterior (R) and interior (r) radii. Breadth (δ) is the width of the annulus. “ So I just have to figure out how to use the online calculator.
The ratio of lucent annulus to dense desmosome is not too apparent when there is no tight junction visible right next to it. But that said, I can see a slight increase in plasmalemmal density for a short distance (same type of annulus arrangement as if there were a tight junction there) on each end of the desomosome cross section.
Junction function (haha)
One cant work on desmosomes and see a reference like this and not laugh. Sometimes scientists do have a sense of humor — not often enough.
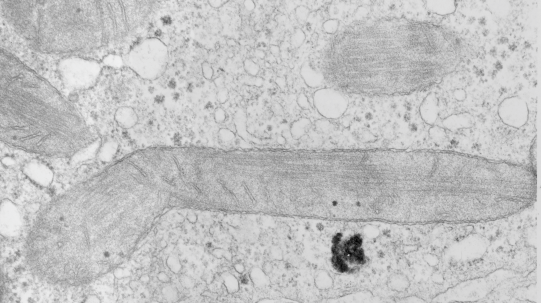
Hepatocyte: rhesus monkey, tubular cristi, mitochondria
Begin here: “empty annulus” outer ring around the desmosomal protein core”
How funny this is, the perimeter of a desmosome with this very precise and quite abrupt “beginning”. It seems that adjacent cells (with a concentric area of pretty well adhered plasmalemmae (two adjacent cells involved)) make the abrupt 45 degree bend just to (coincident with?) the widened intercellular space at the core of the desmosome to its typical width of about 25nm (up from the space between two cells at its closest… more around 17nm).
What is most interesting is what signals the 45 degree spread (bend) on the plasmalemma just before the desmosomal disk, and why is it so electron lucent? or free or proteins?. This is a “free” zone before the electron dense desmosomal complex proteins that make their junctional adherent weld of about 150nm. the width of the annulus. The area of the annulus would vary depending upon where in the spot desmosome the section happened to fall, and a cut in the annulus completely would just look like a wider two-cell junction, showing no desmosome at all. Whether the width of the annulus around the desmosome is consistent from one species, or even one cell type to another is likely not known. I will google to see.
Stub tail monkey hepatocyte desomosomal-mitochondrial tether: more
There are several nice things about this micrograph of a desmosomal mitochondrial tether (i.e. tethered via the intermediate filaments of the desmosome). One nice thing is the very rigid, and parallel membrane-leaflet quality of the plasmalemma where there is the desmosomal protein complex, and it also exhibits a very even and well defined separation of the outer and inner leafelets of the plasmalemma (at least this is what i think is shown in the micrograph below). I have marked the two leaflets on the cell membranes on the adjacent cells with black lines. There are obvious periodicities within the inner and outer membrane leaflets too… not withstanding the density of the lead citrate and uranyl acetate stains and the quality of fixation) which seems to present overall patterning in the entire micrograph. I have marked the extracellular lines (which are not likely to be random as they appear, but more likely to be organized attached wishbone shapes (mirrored vertically and copied horizontally) shown in an earlier post but the lines here have been drown over actual densities in the extracellular (intercellular) space of the desmosome. Green blobs mark periodicities within the two leaflets of the plasmalemma, and the pink blobs mark the densities which are extracellular but likely adjacent, maybe even part of (like the cadherins in desmosomes) those proteins. (green blobs are INTERcellular, the pink blobs are within the plasmalemma (transmembrane). Electron micrograph on left is unretouched, on right, contrast and brightness enhanced and vector overlays show densities which can be matched to image on the left. This stub-tail monkey hepatocyte came from an animal that received two test doses of perfluorodecalin (a very small amount) a year prior, which in my experience has nothing to do with these tethers. Shown immediately below the outer leaflet of the plasmalemma is what appears to be a regular compartmentalization (shown as rectangles) likely representing a highly structure arrangement of plakophilin and plakglobin and maybe some desmoplakin and a bit of vimentin. The number of densities (green or pink, and likely connected parts of the cadherin dimers, turn out in this micrograph to be about 9 nm… a little close than found in mouse micrographs so far. 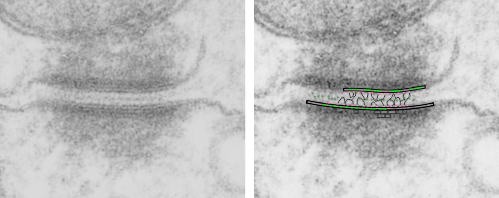
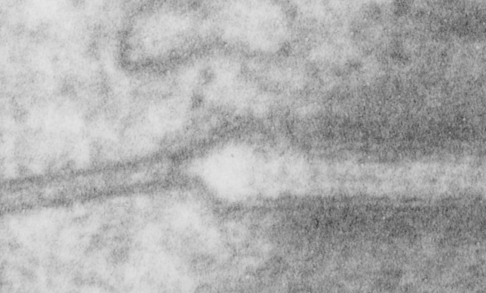
Stub tail monkey hepatocyte desomosomal-mitochondrial tether
These tetherings between desmosomes and mitochondria are pretty plentiful. Here is another example of one such from a hepatocyte from a stub tail monkey. This one shows some very distinct banding…. a little more demarcated than liver, I wonder if this is just opportune sectioning or some molecular difference in the components. Enlargement of the desmosome below top image.
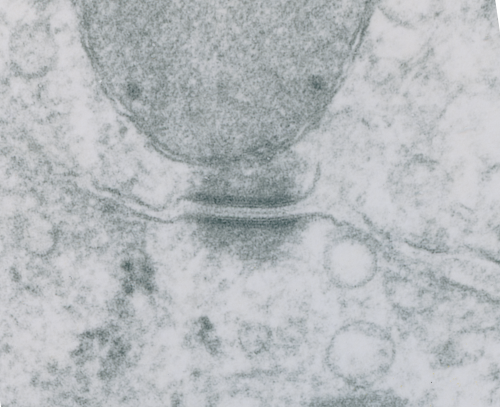
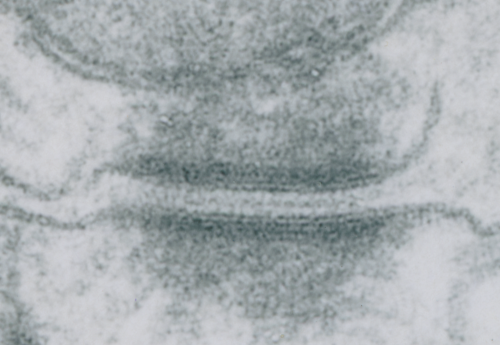 I looks in this image like the plasmalemma of each cell has a very rigid separation of the two leafelets.
I looks in this image like the plasmalemma of each cell has a very rigid separation of the two leafelets.
Desmosomes and mitochondria tethered together
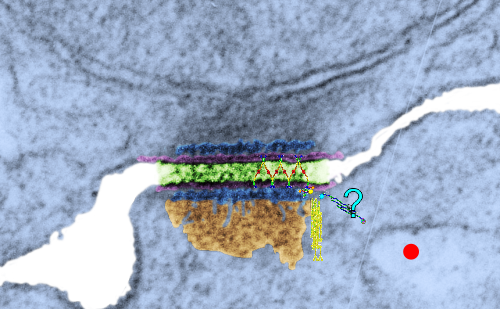
Looking still for some molecular models which fit the images I see for desmosomal mitochondrial junctions (adjacent structures tethered by intermediate filaments). These junctions are so prevalent that I cannot understand why they are not part of the everyday histology lessons. This particular mitochondrion and desmosome are from a mouse liver (control for other studies) . The sectioning and orientation are no too bad though it would be nice to have even more detail (haha as much detail as one might get from electron tomography, which isnt going to happen for me so i will do the best with what i have).
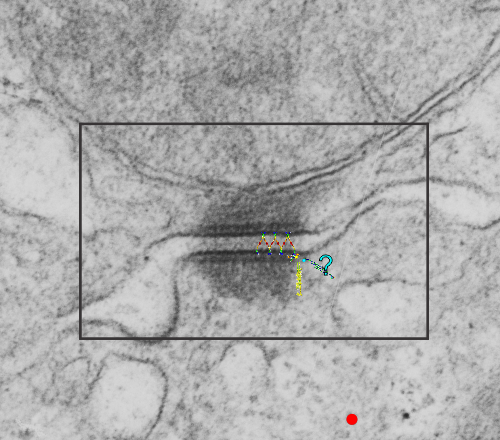
The molecular models of desmosomal proteins, which are pretty widely published, may have relevance to the structures here. The first think I am pretty convinced about is the stretchy hair-pin zigzag like connection of the cadherin molecules within the center of the desomosomal structure, and the little periodicity that is apparent (10-14nm spacing gets calculated from this image). I found a nm marker only on the molecular model for intermediate filaments which was 5nm (a very helpful marker indeed) and it comes close to confirming what I have as the magnification on the micrographs as judged by a 27nm ribosome standard. The proteins overlying the TEM here are just forced to size, as there were no nm markers on those models… and the position of the one large molecule (desmoplakin) is off to the side waiting to be figured out). There are big gaps between what I see with TEM and what the molecules say…. it is fun to try to figure it out.
Upper micrograph, unretouched, also with rectangle for enlarged and pseudocolored image below. The bottom image has the intercellular space colored green, and you can see the cadherin springy wishbones above it, and also the plasmalemma and likely other proteins (plakophilin and plakoglobin) sort of carelessly put in place, and the desmocolin off to the side in an “unknown” placement. Red dots are ribosomes (for size reference).