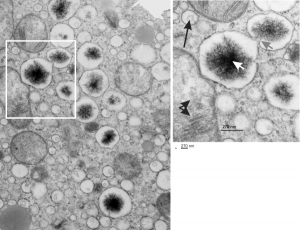
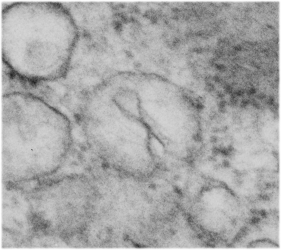
The physical proximity of mitochondria to filaments in the cytoplasm has interested me for a long time. Particularly, those associations between mitochondria and the cytokeratin filaments (tonofilaments, intermediate filaments) on the cytoplasmic side of desmosomes and the associations between mitochondria and nuclear pore filaments (those projecting from the cytoplasmic face of the pore into the cytoplasm). Anchoring plaque proteins and central proteins are pretty nice in this electron micrograph which wikipedia describes thusly: “transmembrane linker proteins, called desmoglein and desmocollin, belong to the cadherin family of cell-adhesion molecules bind to plakoglobin and other proteins in the plaques and extend into the intercellular space, where they interact, forming an interlocking network that binds two cells together”.
I have hundreds of examples of these interactions, the former being more numerous in some cell types, the other more numerous in others. Here is an electron micrograph of one particularly nice association between a mitochondrion and a desmosome. There can be mitochondria on both sides of a desmosome as sectioned in 2D, which begs the question of the 3D aspects of these associations.
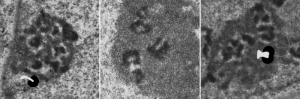
In fact, a single mitochondrion can be associated with more than one desmosome, and can also touch on the cytoplasmic-side-filaments of more than one nuclear pore. M, mitochondrion, arrow, desmosome, bar= 100nm dot is 27 nm by ribosome. Mouse liver female control C4.